Quick Help
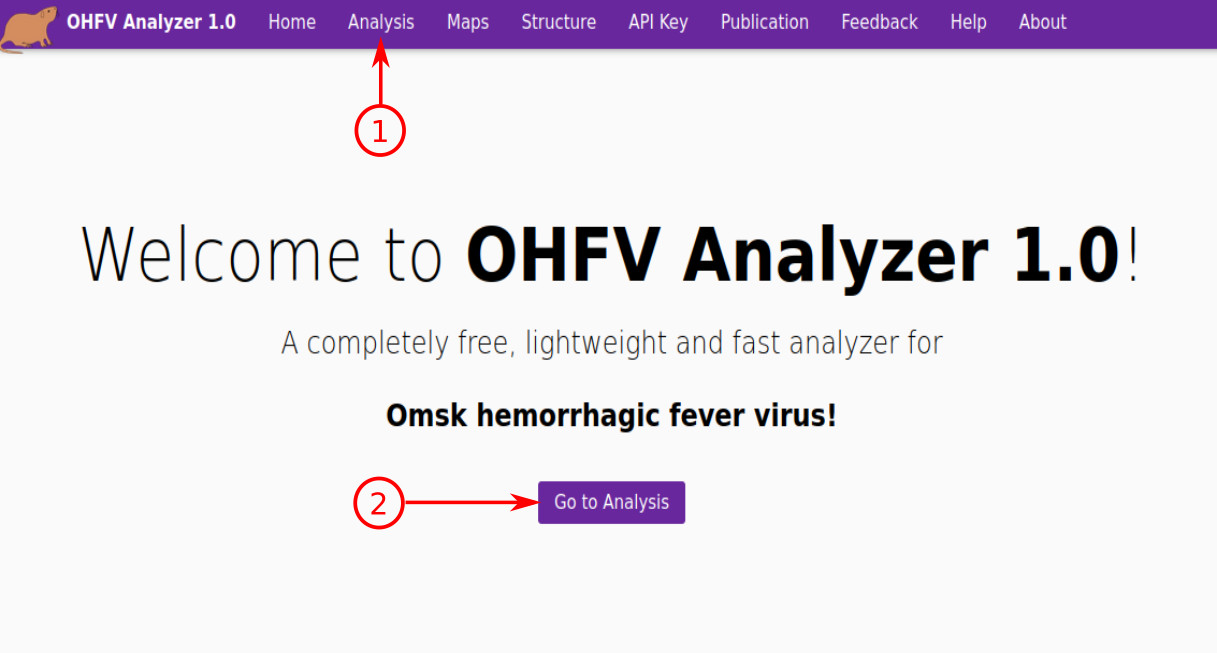
- In order to use the service, press “Analysis” in the navigation menu.
- Also, you can do this by pressing "Go to Analysis" button on the home page.
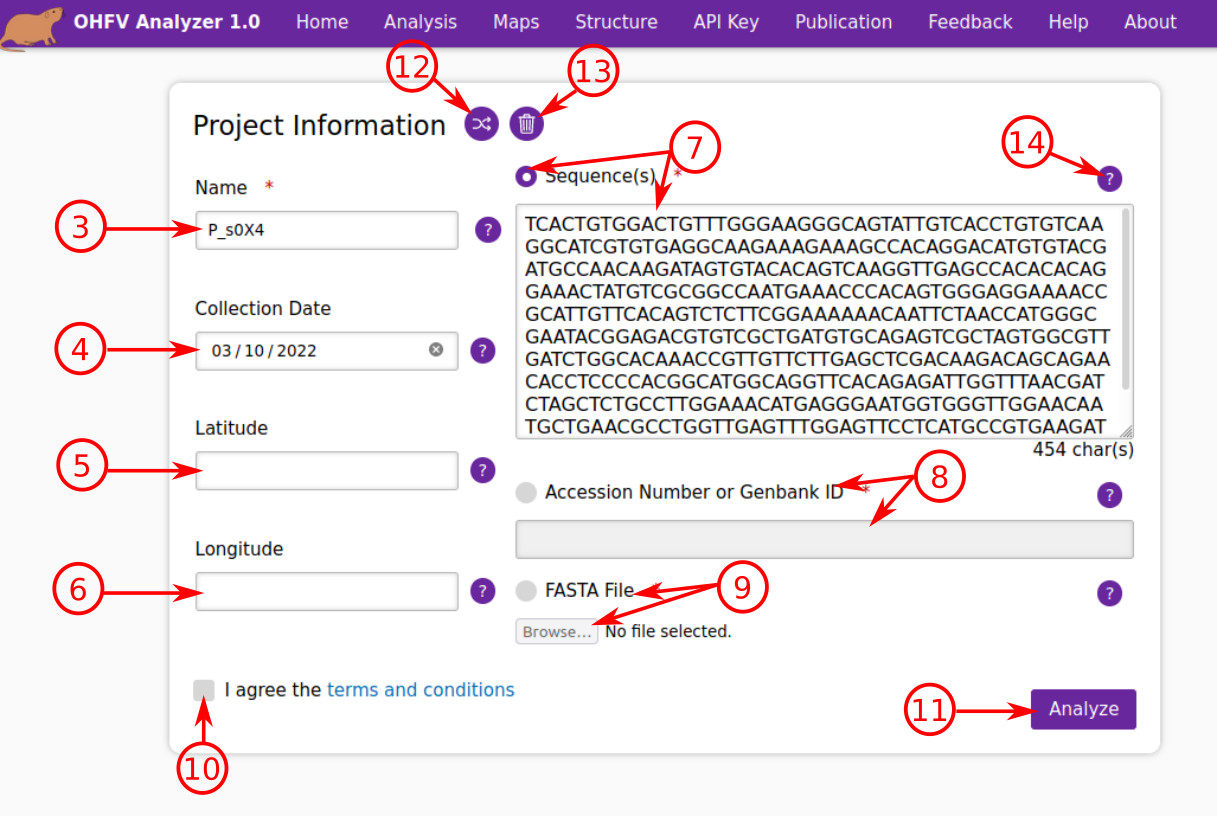
- Enter the name of the project or the query strain name which you wish to analyze. The name must: include 3-16 characters; start with a letter; and contain letters, numbers, underline, and dash characters.
- Enter the collection date for the query strain. (optional)
- Enter the latitude of the collection location for the query strain. (optional)
- Enter the longitude of the collection location for the query strain. (optional)
- Select the radio button "Sequence" and enter the query sequence. The submitted text can include either one or multiple sequences, separated by at least one empty line. Multiple sequences in FASTA-like format are also accepted.
- Select the radio button "Accession Number or Genbank ID" and enter the NCBI accession number or Genbank ID. The submitted text can include either one or multiple IDs/numbers, separated by at least one white space.
- Select the radio button "Upload file" and click on the "Browse..." button in order to upload a local file in FASTA (.fasta or .fas) format for analysis.
- You must accept the terms and conditions to use the service. Press the “terms and conditions” to read its content.
- After pressing this button, the system will check the query information in the form. If there is any error, the system immediately reports it. Otherwise, it will start the pre-processing and then analysis.
- If you want to simply check how the system works, you can press this button to generate a randomly selected sample.
- This button clears the form.
- There is more detailed information about each field, which you can read by hovering the mouse pointer on the question marks.
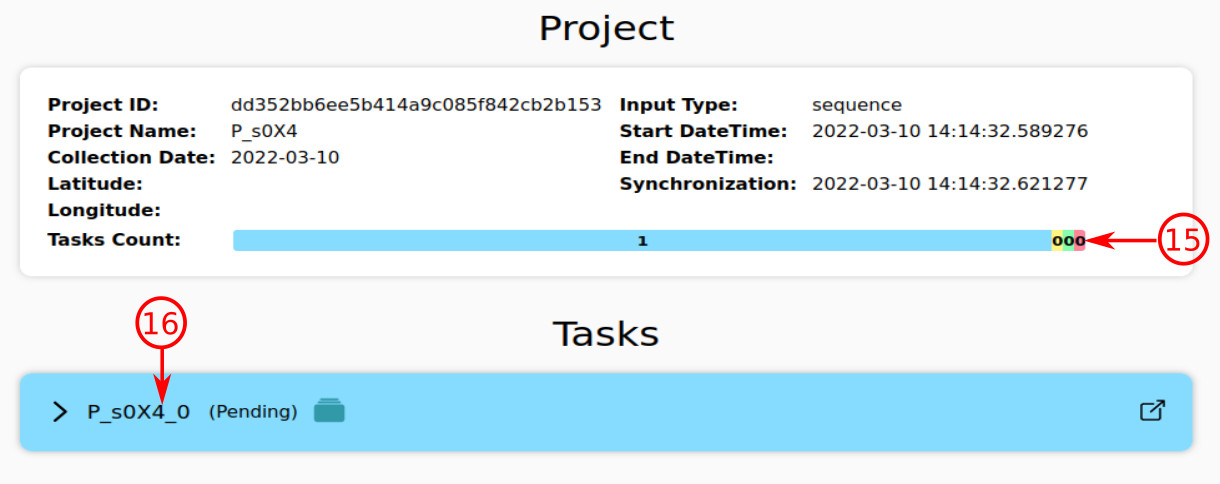
By requesting an analysis, the system will redirect you to the project (monitoring) page, where you can check the status of the submitted task(s).
- This stacked bar provides an overview of the status of all tasks inside the current project. Each color represents a specific status (blue:pending, yellow:runnig, green:succesfully done, red:terminated with error). The content of this page is automatically being refreshed until the platform finishes the tasks either with error or success results.
- Each strain of the query is an individual task with its own status. For example, the current task is "P_uYQZ_0", which has the pending status (blue color).
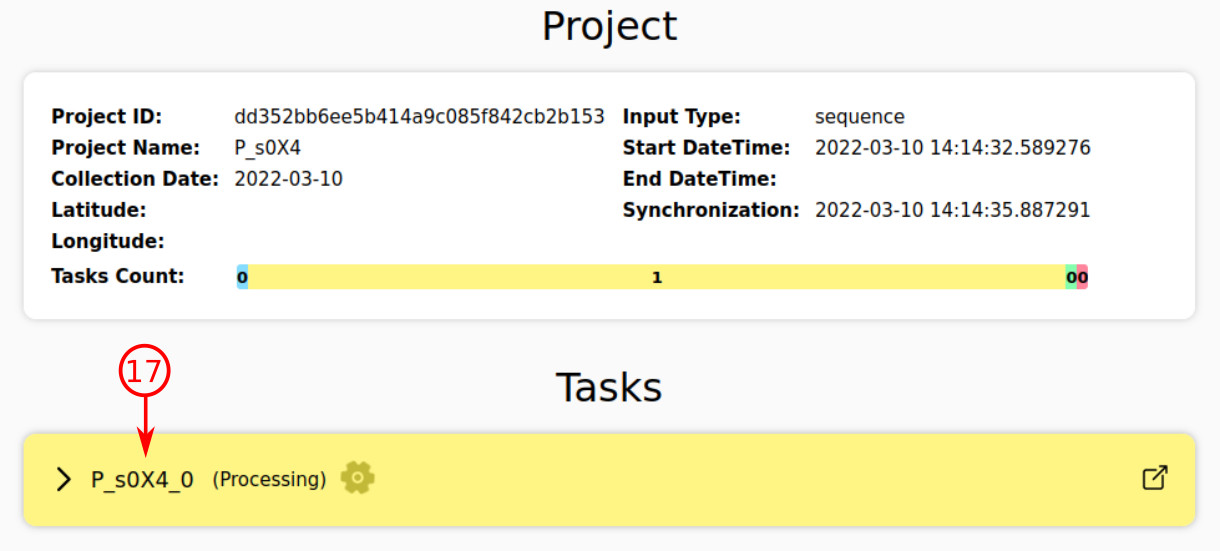
- The color is an indicator of task status; here the color is yellow, so the task is being processed and analyzed.
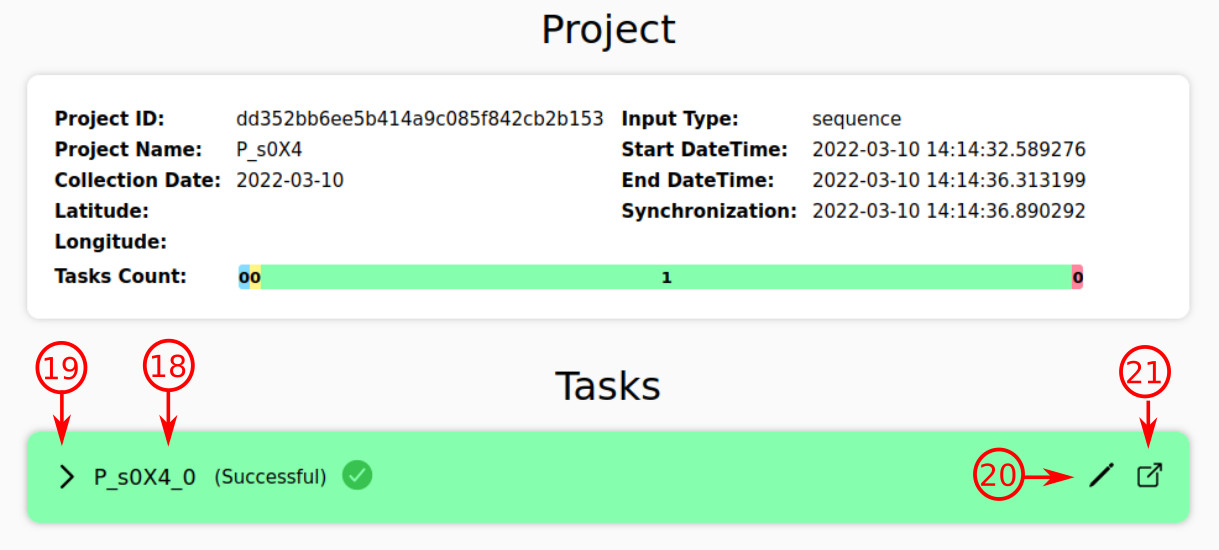
- At this point, the task is successfully finished, and the report is generated.
- By pressing this arrow icon, the green box is expanded to show the brief report.
- As soon as the analysis is carried out, you will be able to rename the task. The new name will be stored on the server and applied in the final report.
- No matter the status of a task, you can refer to the task page, where the final report will be presented, by pressing this icon. An alternative is suggested in the following image (№ 23).
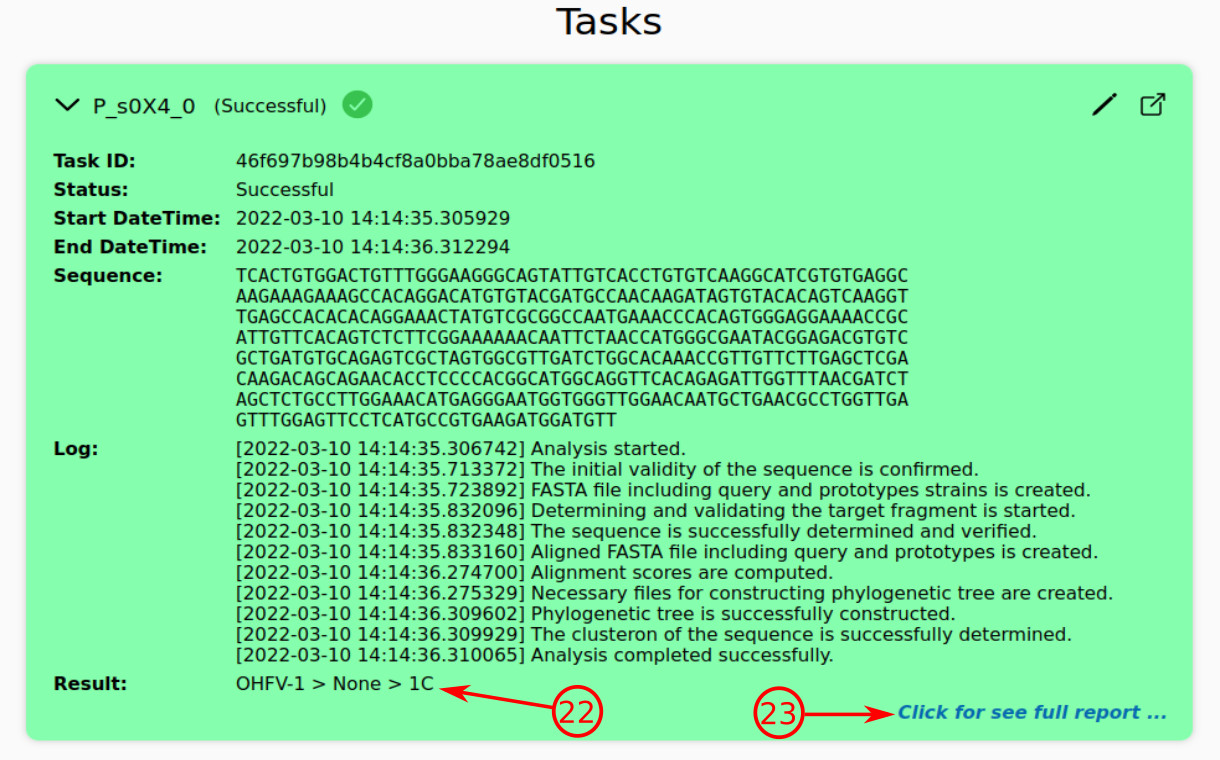
- The brief report presents the main results, including subtype, phylogenetic lineage, and clusteron, respectively, if the task reaches the successful status.
- By clicking this, you can see the full report on the task page.
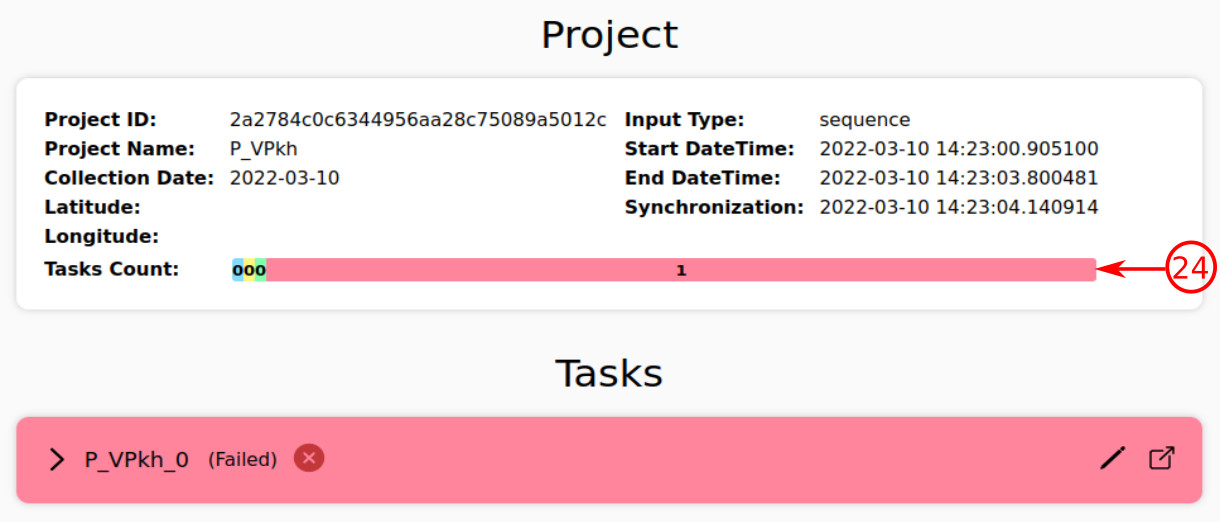
- The platform may encounter some unexpected situations during the analysis process of a task, due to which an error will be raised. The red color represents the existence of error for a task.
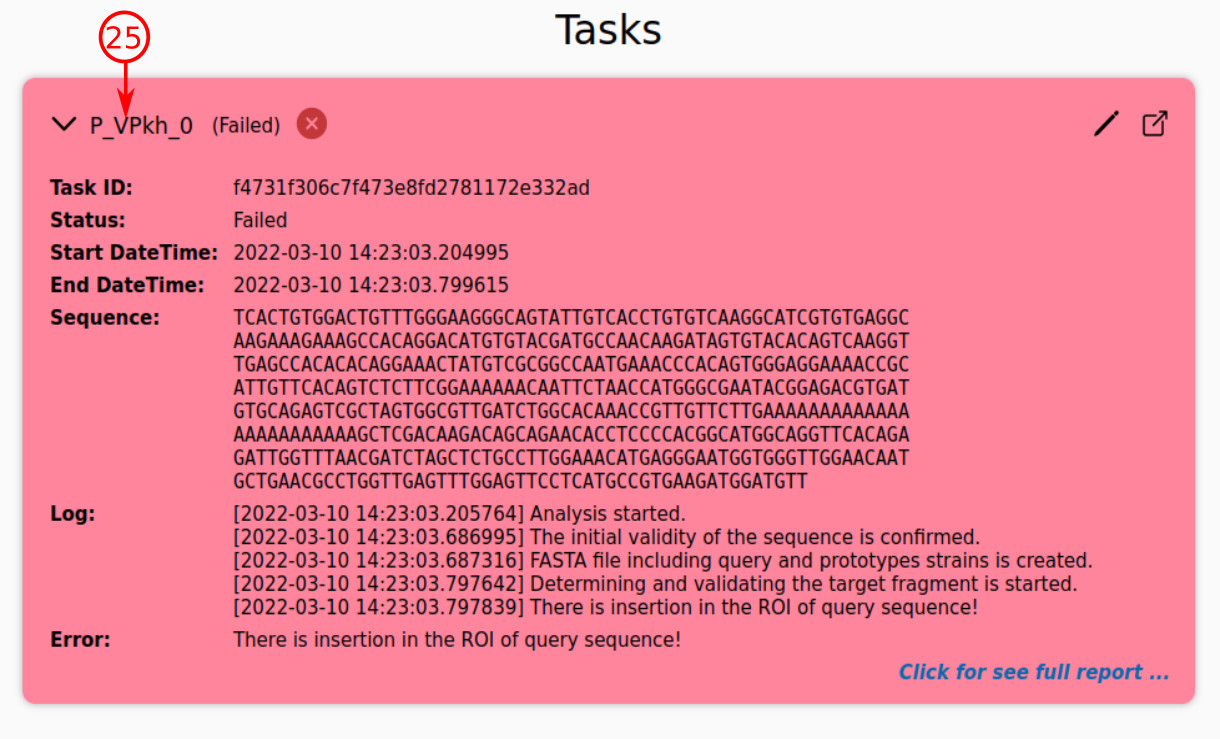
- As you can see, such a task is represented as a failed task. In this case, the user will be informed about the reason for error.
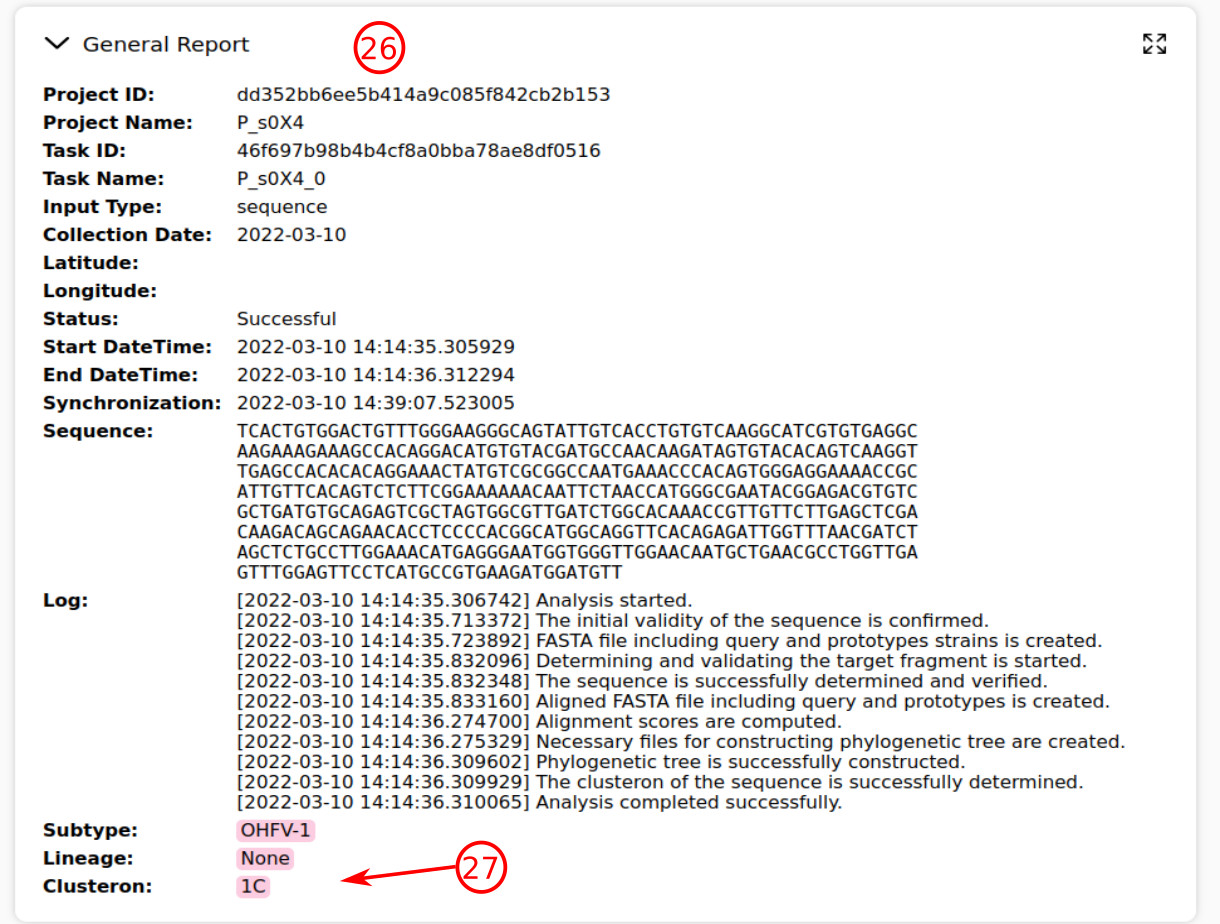
- On the task page, this section shows the general information of the query, system log, and the main results of analysis.
- These are the query subtype, phylogenetic lineage, and the associated clusteron for the query, determined as a result of analysis. Note that phylogenetic lineage is currently considered only for the "Siberian" subtype.
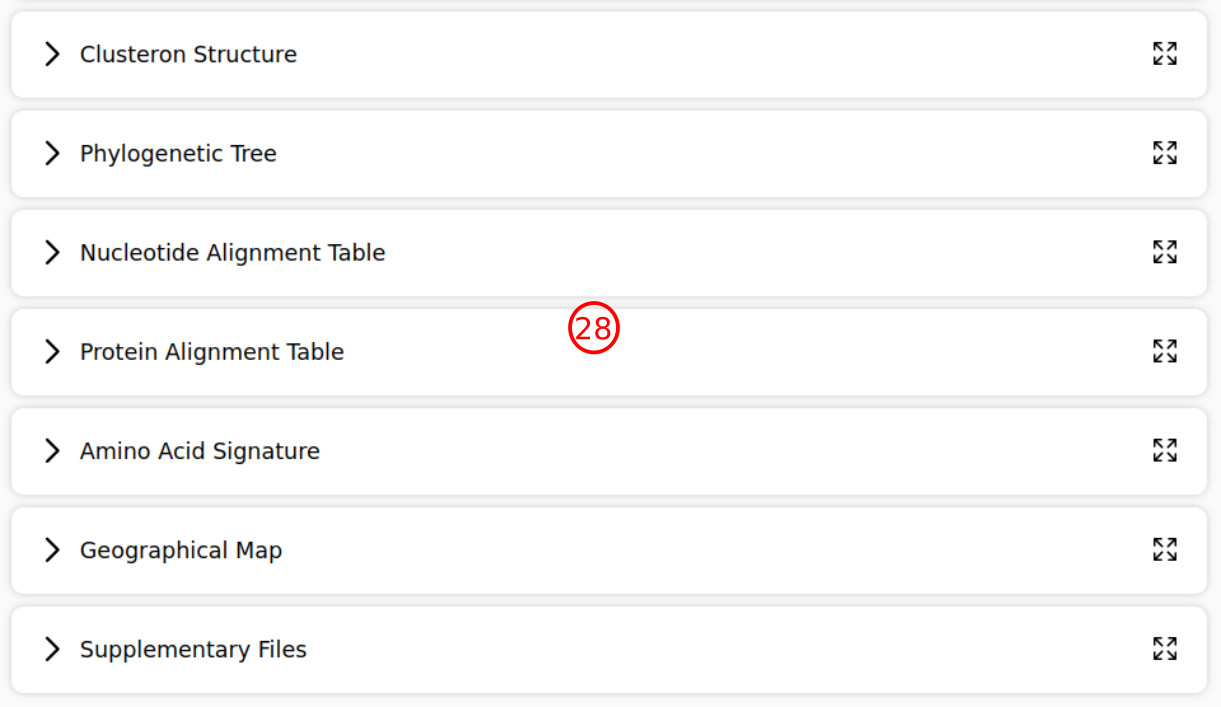
- These are additional reports for the query, which are collapsed by default due to the simplicity of navigation. You can press the left arrow to expand each section.
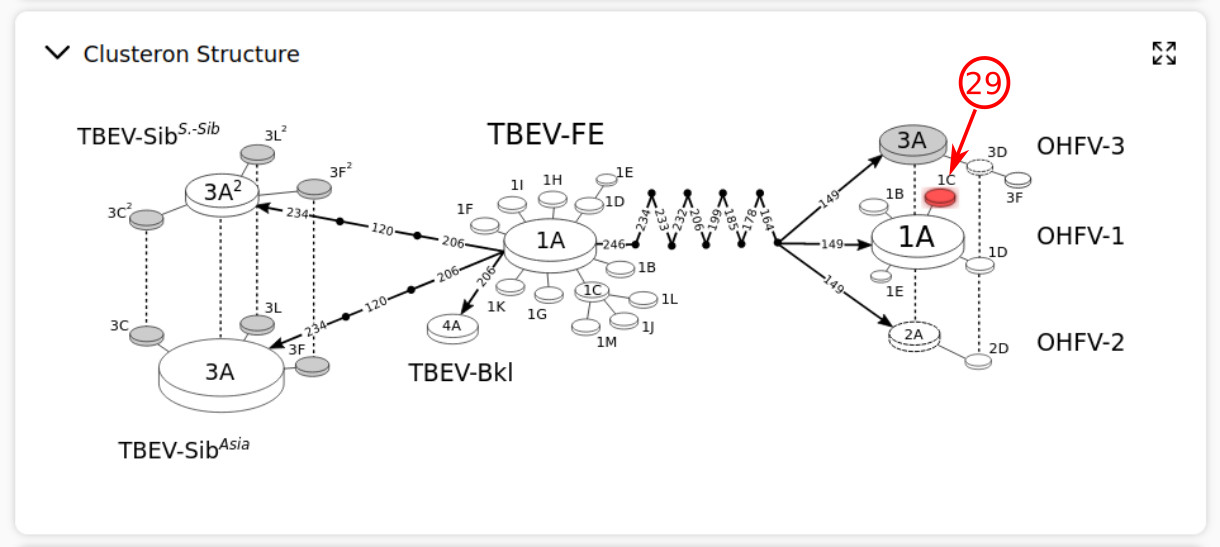
- By expanding the clusteron structure section, the clusteron of the current query is blinking in red color if it is something else than "unique". Note that a unique is usually a new strain that can not be located in the current clusteron structure. More unique strains with similar genetic patterns lead to forming a new clusteron.
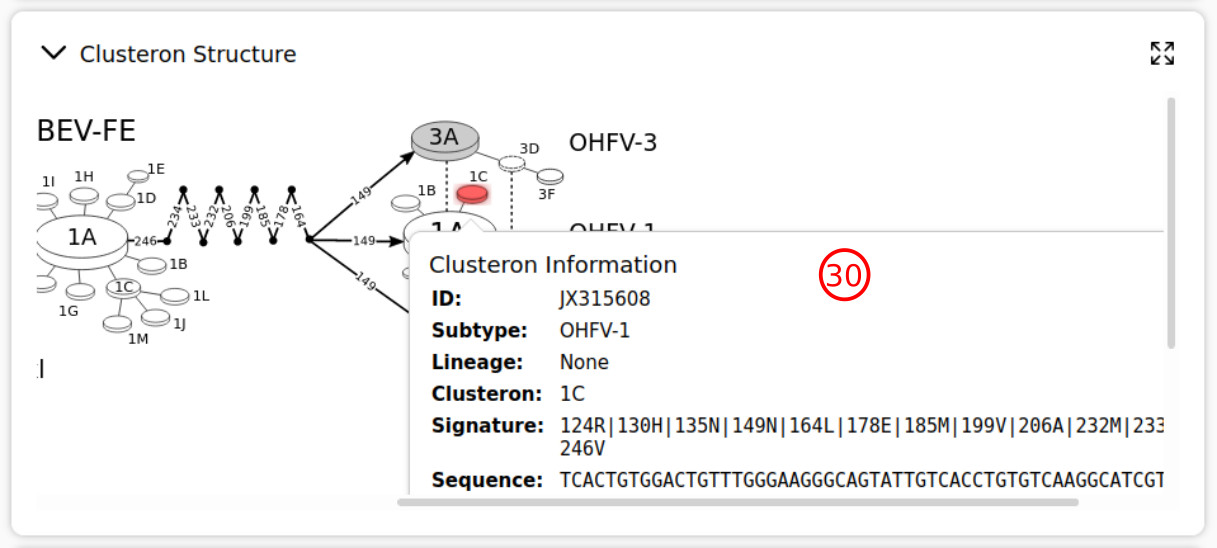
- The profile of each clusteron can be accessed by pressing its node in the graph of clusteron structure.
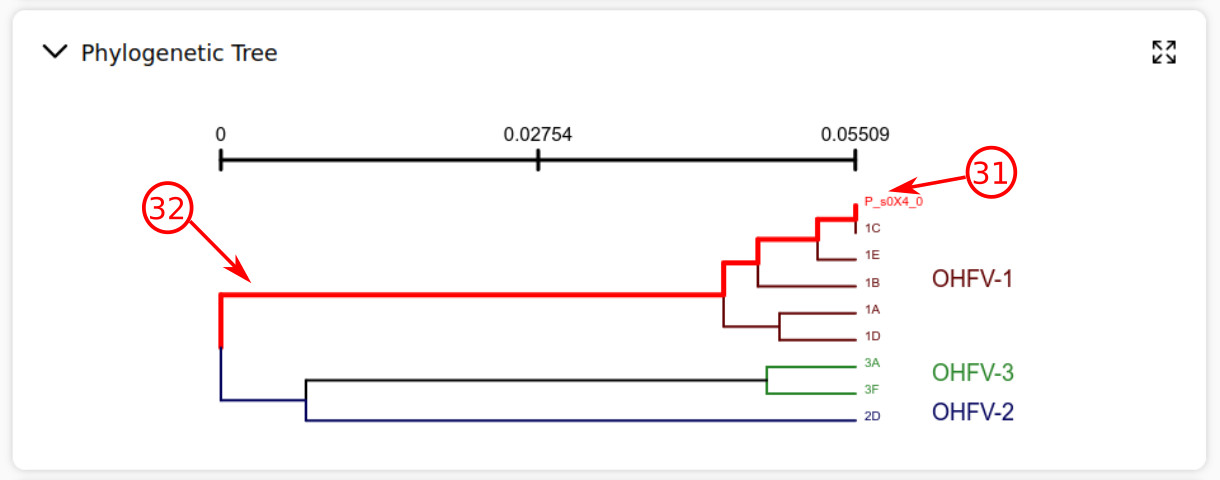
- The most top taxon in the phylogenetic tree belongs to the task strain.
- The task strain path to the root is highlighted in red color.
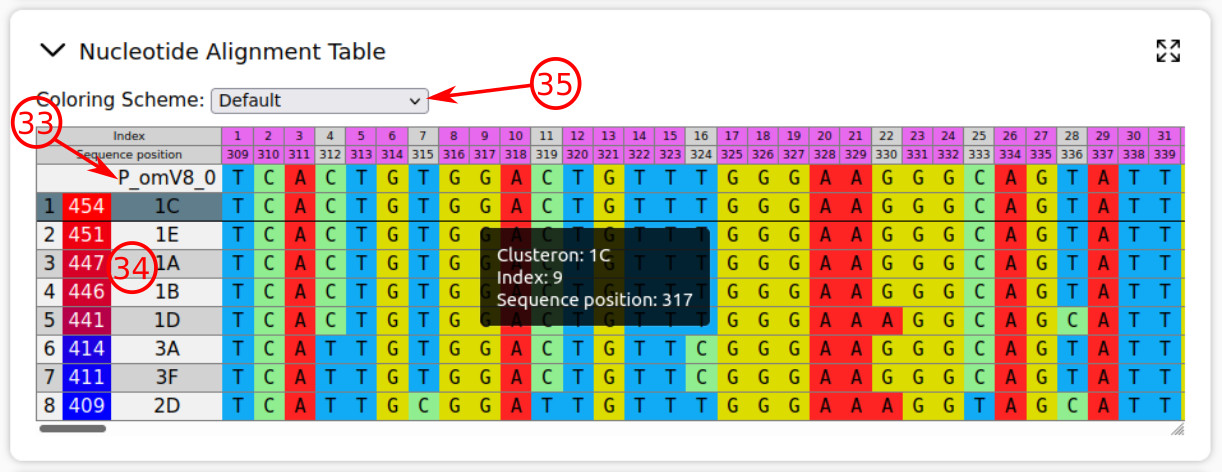
- This is the task query. The current table presents the results of alignment for nucleotide sequences of clusterons and query.
- The clusterons are arranged from high to low degree of similarity, based on the pairwise alignment scores (between query and clusteron sequences). By hovering the mouse pointer on a cell, the information about the cell will be popped up.
- For better visualization of alignment results, several color schemes are provided.
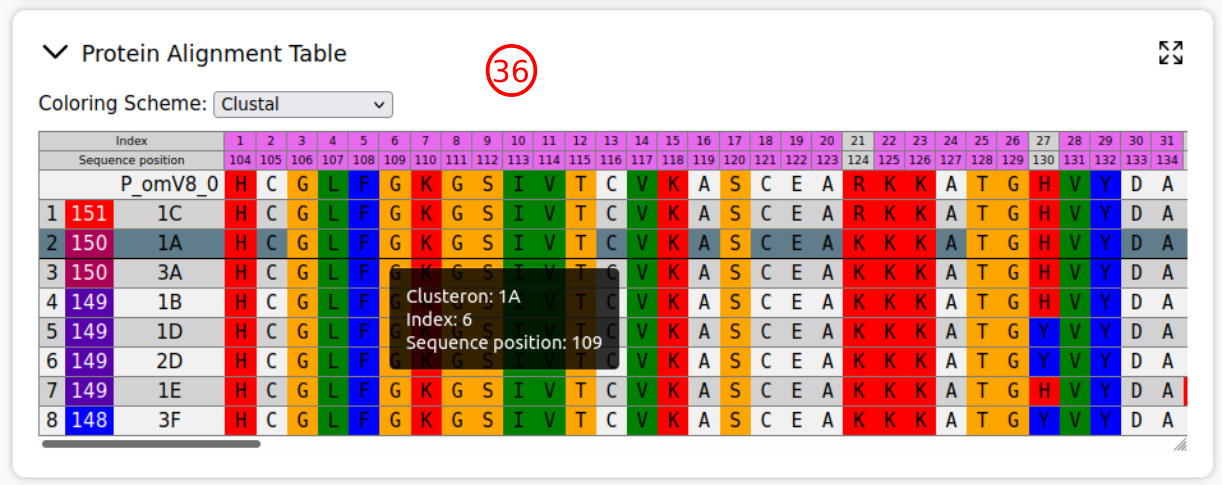
- The protein alignment table also is provided and includes protein sequences of the query and clusterons (prototype). The sequence position indicates the position relative to OHFV/TBEV glycoprotein E. The indices of conserved positions are highlighted in violet. Also, note that the pairwise alignment scores are customized by a gradian color map.
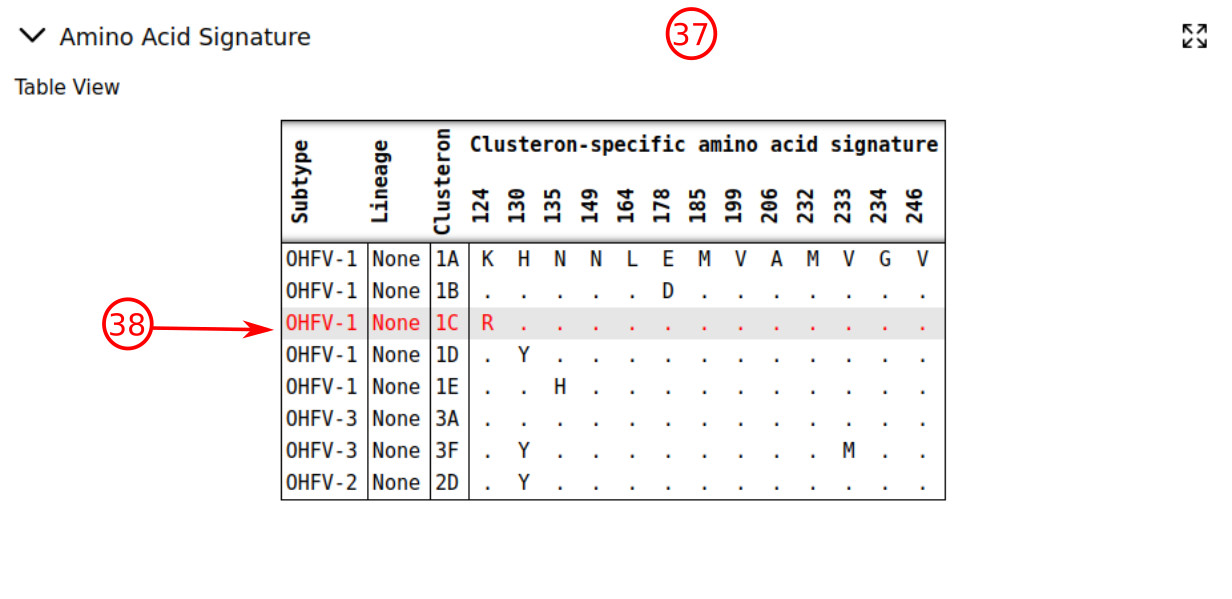
- Clusteron-specific amino acid signature for the clusteron structure can be accessed via the section "Amino Acid Signature".
- The signature of the query is highlighted in red if it is something else than "unique". Moreover, the signature of the assigned clusteron to the query is visualized on the surface of E-protein.
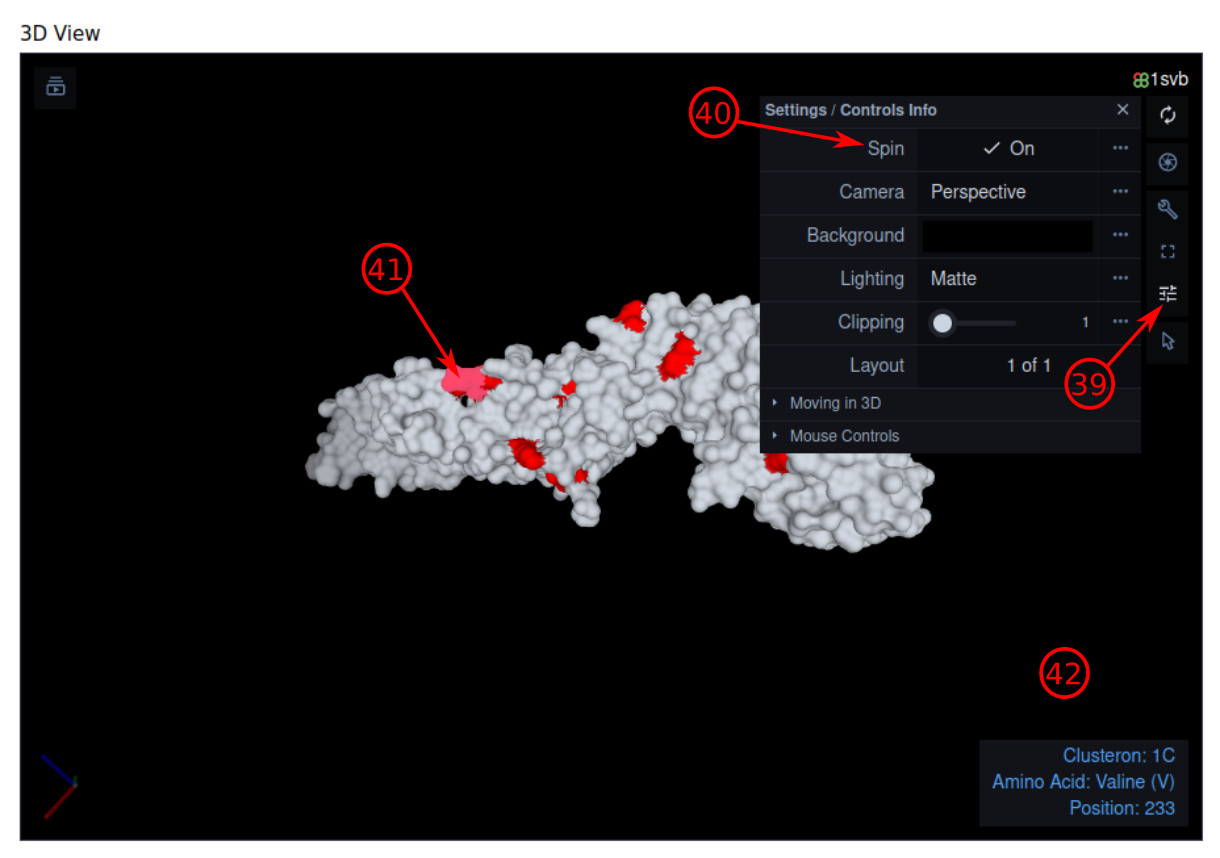
- The 3D visualization of the amino acid signature can be customized by this sidebar menu, e.g., the spinning of 3D geometry can be activated/deactivated. Note that the positions within the signature are highlighted in red on the surface of the protein.
- Toggle "Spin" attribute to on/off the spinning.
- Hover the mouse pointer on a specific site to see the details.
- The information of the hovered site will be shown inside this box.
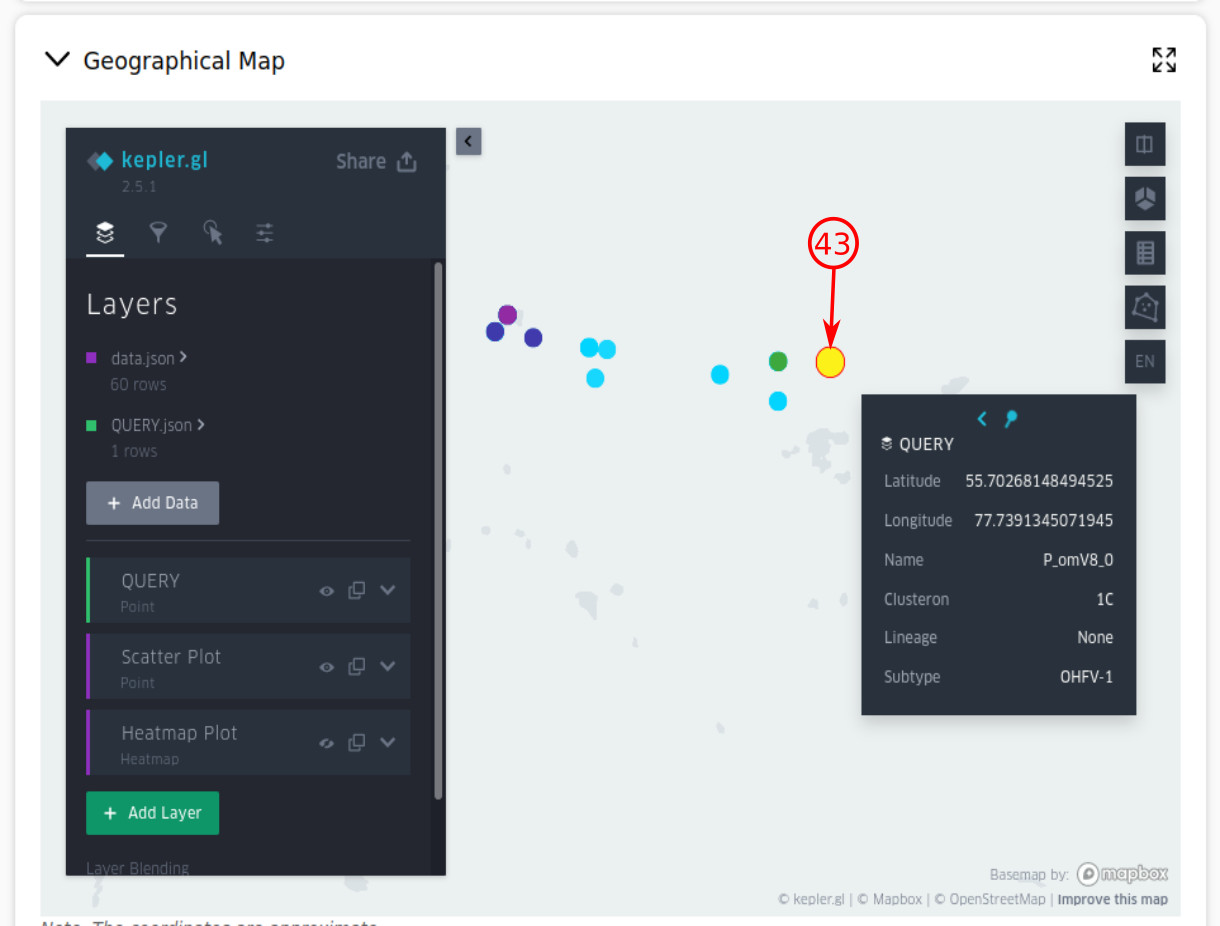
- By providing the latitude and longitude of the query, the geographical map section will be generated inside the task page. So your query will be represented in a red-bordered white-filled circle, on which you can click to see the query information.
- All generated files associated with the query are available via section "Supplementary Files".
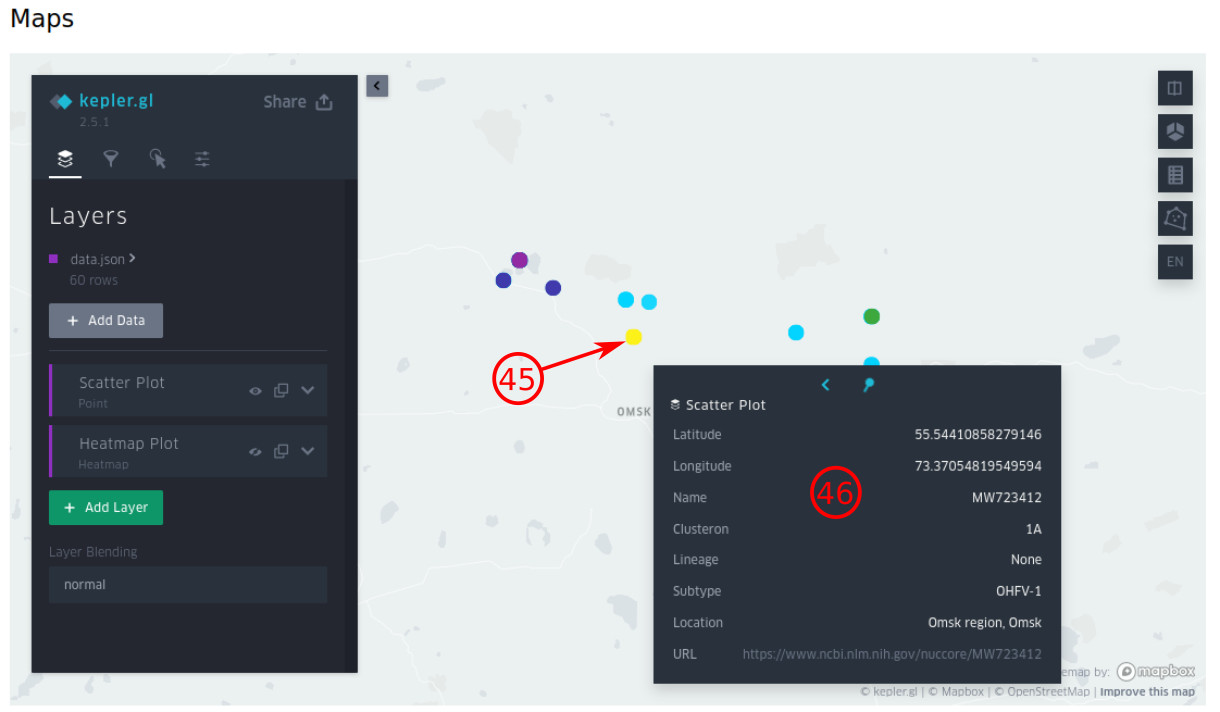
- By pressing "Maps" on the top navigation bar, the scatter plot will be shown by default. So each strain is represented as a point and colored based on its clusteron, on which you can click to see its information.
- The exact latitude and longitude, its NCBI accession number or Genbank ID, clusteron, phylogenetic lineage, and subtype are shown on the popup. Also, you can see more detailed information by click on the provided URL.
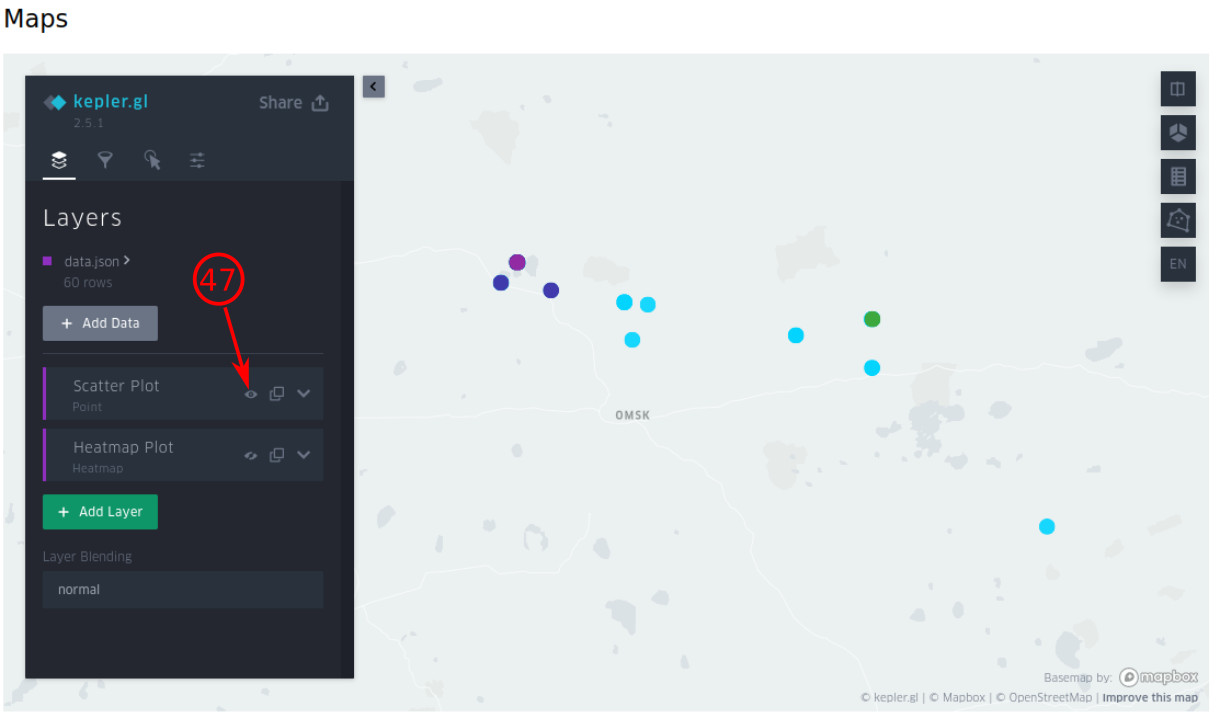
- You can hide/show each layer (or visualization type) by clicking on this eye icon.
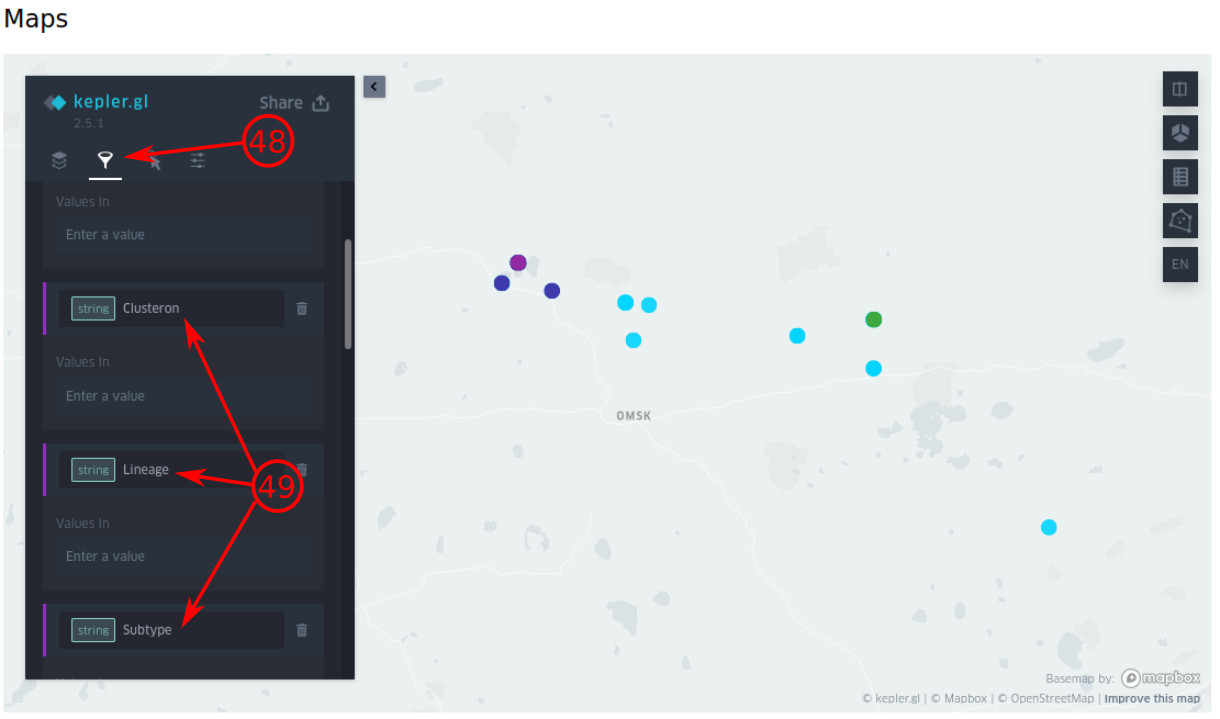
- By clicking this icon, you can filter the visualized objects, e.g., based on clusteron/phylogenetic lineage/clusteron.
- Some filters are provided here by default. You can select the filter by clicking it. (see the next image for filtering the specific objects)
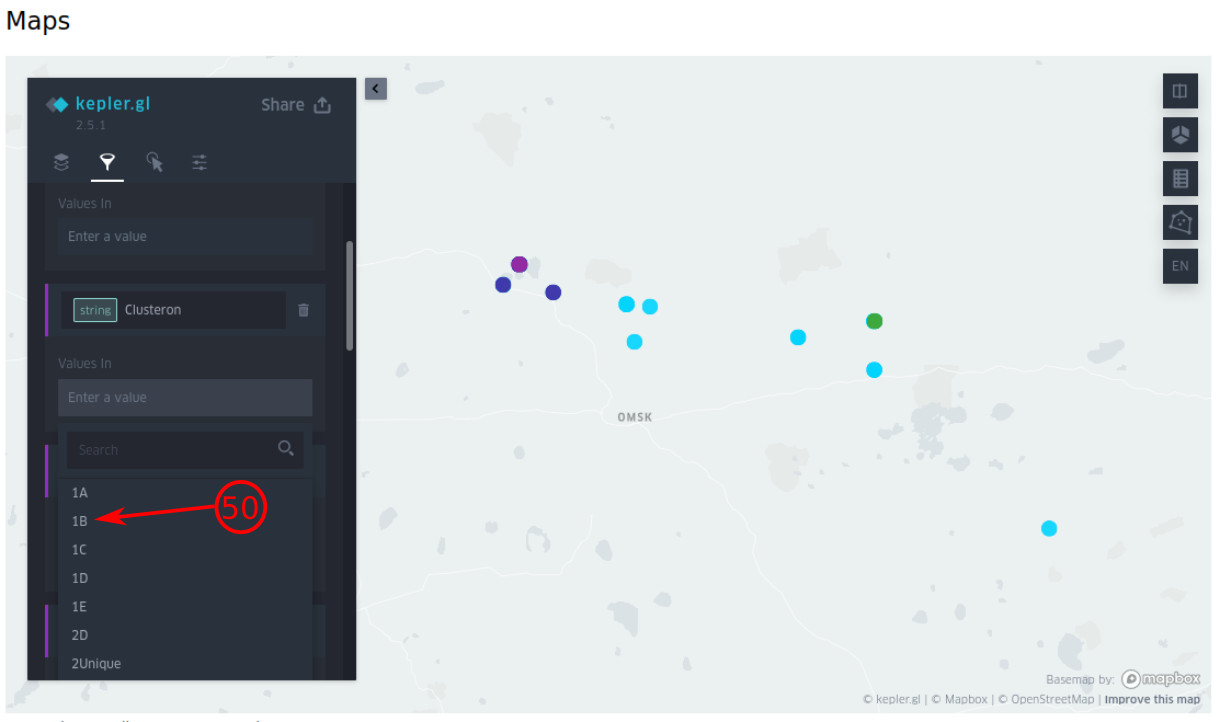
- By selecting the value box, the associated values are automatically presented. Select the value(s) to filter and visualize them on the map
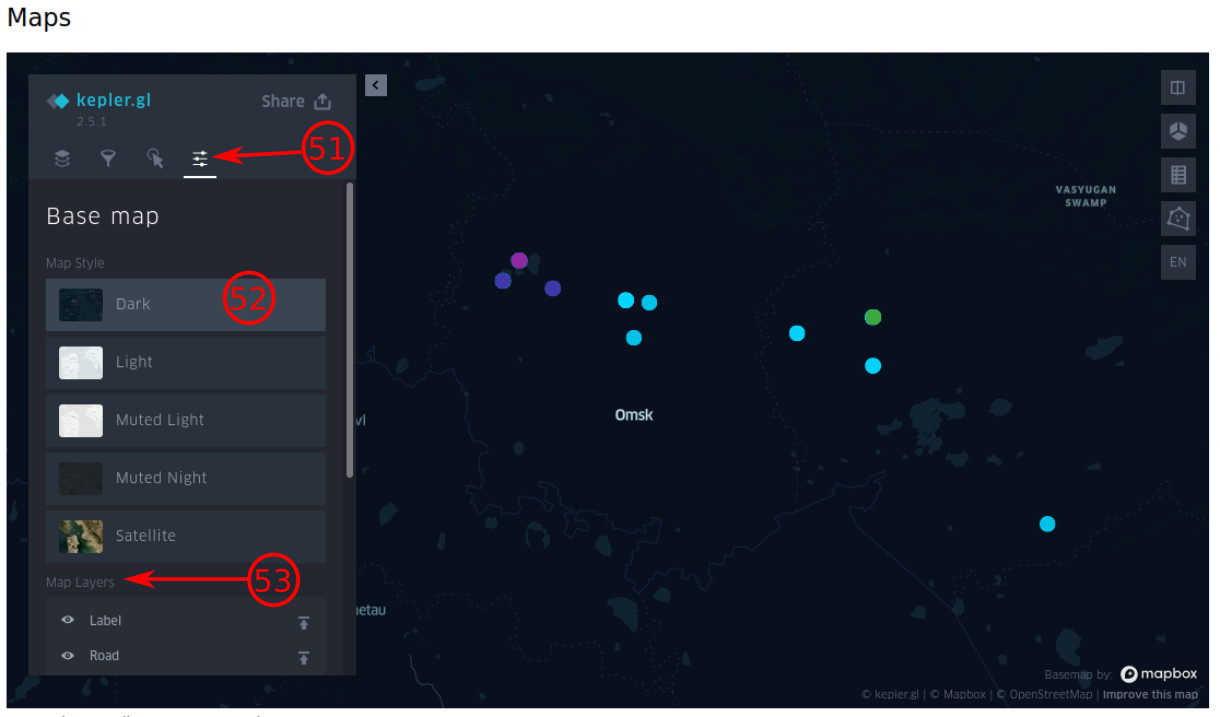
- Click on this icon to refer to the "Base map" section.
- You can customize the style of the map.
- The map layers can be modified in this subsection.
- A visualization type (scatter/heatmap) can also be customized by pressing this arrow.
- As an example, for the heatmap layer, you can adjust the radius by changing the slider value.
- The platform is equipped with programmatic access via API. The API key can be requested by pressing "API Key" in the top navigation bar. Note that the API key will be sent to your Email by our team. There is a 200-queries-per-day limitation. More information about the API is presented in the following section.
API Help
-
To start a new project, you should trigger an HTTP request to
/api/analysiswith the following parameters:key: Your valid API key. (required)name: The name of your project. (required)collection_date: The collection date of query strains. (optional)latitude: The longitude of the collection location. (optional)longitude: The latitude of the collection location. (optional)sequence: The sequence of the query. Note that only one sequence per query is accepted. (required)
Note: The method that is used for HTTP requests must be set toGET.
This is a simple example:
If there are not any errors, the response will be something like this:https://ohfv.viroinformatics.com/api/analysis?key=YOUR_API_KEY&name=YOUR_PROJECT_NAME&sequence=TCACTGTGGACTGTTTGGGAAGGGCAGTATTGTCACCTGTGTCAAGGCATCGTGTGAGGCAAAAAAGAAAGCCACAGGACACGTGTACGACGCTAACAAGATAGTGTACACAGTCAAGGTTGAGCCACATACAGGAAACTATGTCGCGGCCAATGAAACCCACAGTGGGAGGAAAACCGCATTGTTCACAGTCTCTTCGGAAAAAACAATTCTAACCATGGGCGATTACGGGGACGTGTCGCTGATGTGCAGAGTCGCTAGTGGCGTCGATCTGGCACAAACCGTTGTTCTTGAGCTCGACAAGACAGCAGAACACCTCCCCACGGCATGGCAGGTTCACAGAGATTGGTTTAACGATCTAGCTCTGCCTTGGAAACATGAGGGAATGGTGGGTTGGAACAATGCTGAACGCCTGGTTGAGTTTGGAGTTCCTCATGCCGTGAAGATGGATGTT
You can use the field{ "pid": "05f5b4d4804f47318907087827f7eb7f", "success": true, "tid": "50ec97de93b64000a47a9de295768668", "urls": { "api": { "project": "https://ohfv.viroinformatics.com/api/projects?key=2b16dd08e0f54a06b12c72c775772b09&id=05f5b4d4804f47318907087827f7eb7f", "task": "https://ohfv.viroinformatics.com/api/tasks?key=2b16dd08e0f54a06b12c72c775772b09&pid=05f5b4d4804f47318907087827f7eb7f&tid=50ec97de93b64000a47a9de295768668" }, "ui": { "project": "https://ohfv.viroinformatics.com/projects?id=05f5b4d4804f47318907087827f7eb7f", "task": "https://ohfv.viroinformatics.com/tasks?pid=05f5b4d4804f47318907087827f7eb7f&tid=50ec97de93b64000a47a9de295768668" } } }urlsto access the details of the project or its task through either API or web browser.
Thesuccessproperty indicates that whether the project is created or there are some errors. -
To access the details of a project, you should trigger an HTTP request to
/api/projectswith these parameters:key: Your valid API key. (required)id: The ID of the project. (required)
And the response for a project with a successful task will be something like this:https://ohfv.viroinformatics.com/api/projects?key=YOUR_API_KEY&id=05f5b4d4804f47318907087827f7eb7f{ "_id": "05f5b4d4804f47318907087827f7eb7f", "collection_date": "", "datetime": { "end": "2022-03-09 14:11:04.607314", "start": "2022-03-09 14:10:53.348658" }, "ip": "127.0.0.1", "latitude": "", "longitude": "", "name": "Test", "tasks": [ { "datetime": { "end": "2022-03-09 14:11:04.603747", "start": "2022-03-09 14:10:59.000907" }, "id": "50ec97de93b64000a47a9de295768668", "log": [ [ "message", "2022-03-09 14:10:59.034277", "Analysis started." ], [ "message", "2022-03-09 14:10:59.066561", "The initial validity of the sequence is confirmed." ], [ "message", "2022-03-09 14:10:59.144560", "FASTA file including query and prototypes strains is created." ], [ "message", "2022-03-09 14:10:59.425692", "Determining and validating the target fragment is started." ], [ "message", "2022-03-09 14:10:59.426327", "The sequence is successfully determined and verified." ], [ "message", "2022-03-09 14:10:59.428535", "Aligned FASTA file including query and prototypes is created." ], [ "message", "2022-03-09 14:11:04.448737", "Alignment scores are computed." ], [ "message", "2022-03-09 14:11:04.449943", "Necessary files for constructing phylogenetic tree are created." ], [ "message", "2022-03-09 14:11:04.596387", "Phylogenetic tree is successfully constructed." ], [ "message", "2022-03-09 14:11:04.597070", "The clusteron of the sequence is successfully determined." ], [ "success", "2022-03-09 14:11:04.597640", "Analysis completed successfully." ] ], "name": "Test", "report": { "aligned": ">Test\nTCACTGTGGACTGTTTGGGAAGGGCAGTATTGTCACCTGTGTCAAGGCATCGTGTGAGGC\nAAAAAAGAAAGCCACAGGACACGTGTACGACGCTAACAAGATAGTGTACACAGTCAAGGT\nTGAGCCACATACAGGAAACTATGTCGCGGCCAATGAAACCCACAGTGGGAGGAAAACCGC\nATTGTTCACAGTCTCTTCGGAAAAAACAATTCTAACCATGGGCGATTACGGGGACGTGTC\nGCTGATGTGCAGAGTCGCTAGTGGCGTCGATCTGGCACAAACCGTTGTTCTTGAGCTCGA\nCAAGACAGCAGAACACCTCCCCACGGCATGGCAGGTTCACAGAGATTGGTTTAACGATCT\nAGCTCTGCCTTGGAAACATGAGGGAATGGTGGGTTGGAACAATGCTGAACGCCTGGTTGA\nGTTTGGAGTTCCTCATGCCGTGAAGATGGATGTT\n>1A\nTCACTGTGGACTGTTTGGGAAGGGCAGTATTGTCACCTGTGTCAAGGCATCGTGTGAGGC\nAAAGAAGAAAGCCACAGGACATGTGTATGACGCTAACAAGATAGTGTACACAGTCAAGGT\nTGAGCCACATACAGGAAACTATGTCGCGGCCAATGAAACCCATAGTGGGAGGAAAACCGC\nATTGTTCACAGTCTCTTCGGAAAAAACAATTCTAACCATGGGCGAATACGGAGACGTGTC\nGCTGATGTGCAGAGTCGCTAGTGGCGTCGATCTGGCACAAACCGTTGTTCTTGAGCTCGA\nCAAGACAGCAGAACACCTCCCCACGGCATGGCAGGTTCACAGAGATTGGTTTAACGATCT\nAGCTCTGCCTTGGAAACATGAGGGAATGGTGGGTTGGAACAATGCTGAACGCCTGGTTGA\nGTTTGGAGTTCCTCATGCCGTGAAGATGGATGTT\n>3A\nTCATTGTGGACTGTTCGGGAAGGGCAGTATTGTTACCTGTGTTAAGGCATCATGTGAGGC\nAAAAAAGAAAGCCACGGGACACGTGTATGATGCAAACAAGATAGTGTACACAGTCAAGGT\nCGAGCCACACACAGGAAACTATGTTGCAGCCAATGAAACCCACAGTGGGAGGAAAACCGC\nGTTATTTACGGTCTCTTCGGAAAAAACCATCTTGACCATGGGTGAATATGGAGACGTGTC\nATTGATGTGCAGAGTTGCCAGCGGCGTTGACTTGGCACAGACCGTTGTTCTTGAGCTCGA\nCAAGACAGCAGAACACCTCCCTACGGCATGGCAGGTTCACAGAGACTGGTTTAATGACCT\nGGCTCTGCCATGGAAGCATGAGGGAATGGTGGGTTGGAACAATGCAGAACGCCTGGTTGA\nGTTCGGAGTTCCTCATGCCGTGAAGATGGACGTG\n>1B\nTCACTGTGGACTGTTTGGGAAGGGCAGTATTGTCACCTGTGTCAAGGCATCGTGTGAGGC\nAAAAAAGAAAGCCACAGGACACGTGTACGACGCTAACAAGATAGTGTACACAGTCAAGGT\nTGAGCCACATACAGGAAACTATGTCGCGGCCAATGAAACCCACAGTGGGAGGAAAACCGC\nATTGTTCACAGTCTCTTCGGAAAAAACAATTCTAACCATGGGCGATTACGGGGACGTGTC\nGCTGATGTGCAGAGTCGCTAGTGGCGTCGATCTGGCACAAACCGTTGTTCTTGAGCTCGA\nCAAGACAGCAGAACACCTCCCCACGGCATGGCAGGTTCACAGAGATTGGTTTAACGATCT\nAGCTCTGCCTTGGAAACATGAGGGAATGGTGGGTTGGAACAATGCTGAACGCCTGGTTGA\nGTTTGGAGTTCCTCATGCCGTGAAGATGGATGTT\n>1C\nTCACTGTGGACTGTTTGGGAAGGGCAGTATTGTCACCTGTGTCAAGGCATCGTGTGAGGC\nAAGAAAGAAAGCCACAGGACATGTGTACGATGCCAACAAGATAGTGTACACAGTCAAGGT\nTGAGCCACACACAGGAAACTATGTCGCGGCCAATGAAACCCACAGTGGGAGGAAAACCGC\nATTGTTCACAGTCTCTTCGGAAAAAACAATTCTAACCATGGGCGAATACGGAGACGTGTC\nGCTGATGTGCAGAGTCGCTAGTGGCGTTGATCTGGCACAAACCGTTGTTCTTGAGCTCGA\nCAAGACAGCAGAACACCTCCCCACGGCATGGCAGGTTCACAGAGATTGGTTTAACGATCT\nAGCTCTGCCTTGGAAACATGAGGGAATGGTGGGTTGGAACAATGCTGAACGCCTGGTTGA\nGTTTGGAGTTCCTCATGCCGTGAAGATGGATGTT\n>1D\nTCACTGTGGACTGTTTGGGAAAGGCAGCATTGTTACCTGTGTCAAGGCATCGTGTGAGGC\nAAAGAAGAAAGCCACAGGATATGTGTATGACGCTAACAAGATAGTGTACACAGTCAAGGT\nTGAGCCACATACAGGAAACTATGTCGCGGCCAATGAAACCCATAGTGGGAGAAAAACCGC\nATTGTTCACAGTCTCTTCGGAAAAAACAATTCTAACCATGGGCGAATACGGAGACGTGTC\nGCTGATGTGCAGAGTCGCTAGTGGCGTCGATCTGGCACAAACCGTTGTTCTTGAGCTCGA\nCAAGACAGCAGAACACCTCCCCACGGCATGGCAGGTTCACAGAGATTGGTTTAACGATCT\nAGCTCTGCCTTGGAAACATGAGGGAATGGTGGGTTGGAACAATGCTGAACGCCTGGTTGA\nGTTTGGAGTTCCTCACGCCGTGAAGATGGATGTT\n>2D\nTCATTGCGGATTGTTTGGGAAAGGTAGCATTGTCACCTGTGTCAAGGCATCATGTGAGGC\nAAAAAAGAAAGCCACGGGATATGTTTATGATGCAAACAAGATAGTGTACACGGTCAAGGT\nTGAGCCACACACAGGGAACTATGTCGCAGCCAACGAAACCCACAGTGGGAGGAAAACTGC\nGCTGTTCACGGTCTCCTCGGAAAAAACCATCCTGACCATGGGTGAATATGGAGACGTGTC\nATTGATGTGTAGAGTCGCTAGTGGCGTTGACTTGGCACAGACCGTTGTCCTCGAGCTTGA\nCAAGACAGCAGAACACCTTCCCACAGCATGGCAGGTTCACAGAGATTGGTTTAATGATCT\nGGCTCTGCCATGGAAACATGAGGGAATGGTGGGCTGGAACAATGCAGAACGCCTGGTTGA\nGTTTGGAGTCCCGCATGCTGTGAAGATGGACGTT\n>1E\nTCACTGTGGACTGTTTGGGAAGGGCAGTATTGTCACCTGTGTCAAGGCATCGTGTGAGGC\nAAAAAAGAAAGCCACAGGACATGTGTACGACGCCCACAAGATAGTGTACACAGTCAAGGT\nTGAGCCACACACAGGAAACTATGTCGCGGCCAATGAAACCCACAGTGGGAGGAAAACCGC\nATTGTTCACAGTCTCTTCGGAAAAAACAATTCTAACCATGGGCGAATACGGAGACGTGTC\nGCTGATGTGCAGAGTCGCTAGTGGCGTTGATCTGGCACAAACCGTTGTTCTTGAGCTCGA\nCAAGACAGCAGAACACCTCCCCACGGCATGGCAGGTTCACAGAGATTGGTTTAACGATCT\nAGCTCTGCCTTGGAAACATGAGGGAATGGTGGGTTGGAACAATGCTGAACGCCTGGTTGA\nGTTTGGAGTTCCTCATGCCGTGAAGATGGATGTT\n>3F\nTCATTGTGGACTGTTCGGGAAGGGCAGTATTGTTACCTGTGTTAAGGCATCATGTGAGGC\nAAAAAAGAAAGCCACGGGATACGTGTATGATGCAAACAAGATAGTGTATACAGTCAAGGT\nCGAGCCACACACAGGAAACTATGTTGCAGCCAATGAAACCCACAGTGGGAGGAAAACCGC\nGTTATTCACGGTCTCTTCGGAAAAGACCATCTTGACCATGGGTGAATATGGAGACGTGTC\nATTGATGTGCAGAGTTGCCAGTGGCGTTGACTTGGCACAGACCGTTGTTCTTGAGCTCGA\nCAAGACAGCAGAACACCTCCCTACGGCATGGCAGGTTCACAGAGACTGGTTTAATGACCT\nGGCTCTGCCATGGAAGCATGAGGGAATGATGGGATGGAACAATGCAGAACGCCTGGTTGA\nGTTCGGAGTTCCTCATGCCGTGAAGATGGACGTG\n", "clusteron": "1B", "lineage": "None", "scores_dna": "[[-1, \"Test\", \"TCACTGTGGACTGTTTGGGAAGGGCAGTATTGTCACCTGTGTCAAGGCATCGTGTGAGGCAAAAAAGAAAGCCACAGGACACGTGTACGACGCTAACAAGATAGTGTACACAGTCAAGGTTGAGCCACATACAGGAAACTATGTCGCGGCCAATGAAACCCACAGTGGGAGGAAAACCGCATTGTTCACAGTCTCTTCGGAAAAAACAATTCTAACCATGGGCGATTACGGGGACGTGTCGCTGATGTGCAGAGTCGCTAGTGGCGTCGATCTGGCACAAACCGTTGTTCTTGAGCTCGACAAGACAGCAGAACACCTCCCCACGGCATGGCAGGTTCACAGAGATTGGTTTAACGATCTAGCTCTGCCTTGGAAACATGAGGGAATGGTGGGTTGGAACAATGCTGAACGCCTGGTTGAGTTTGGAGTTCCTCATGCCGTGAAGATGGATGTT\"], [454, \"1B\", \"TCACTGTGGACTGTTTGGGAAGGGCAGTATTGTCACCTGTGTCAAGGCATCGTGTGAGGCAAAAAAGAAAGCCACAGGACACGTGTACGACGCTAACAAGATAGTGTACACAGTCAAGGTTGAGCCACATACAGGAAACTATGTCGCGGCCAATGAAACCCACAGTGGGAGGAAAACCGCATTGTTCACAGTCTCTTCGGAAAAAACAATTCTAACCATGGGCGATTACGGGGACGTGTCGCTGATGTGCAGAGTCGCTAGTGGCGTCGATCTGGCACAAACCGTTGTTCTTGAGCTCGACAAGACAGCAGAACACCTCCCCACGGCATGGCAGGTTCACAGAGATTGGTTTAACGATCTAGCTCTGCCTTGGAAACATGAGGGAATGGTGGGTTGGAACAATGCTGAACGCCTGGTTGAGTTTGGAGTTCCTCATGCCGTGAAGATGGATGTT\"], [448, \"1A\", \"TCACTGTGGACTGTTTGGGAAGGGCAGTATTGTCACCTGTGTCAAGGCATCGTGTGAGGCAAAGAAGAAAGCCACAGGACATGTGTATGACGCTAACAAGATAGTGTACACAGTCAAGGTTGAGCCACATACAGGAAACTATGTCGCGGCCAATGAAACCCATAGTGGGAGGAAAACCGCATTGTTCACAGTCTCTTCGGAAAAAACAATTCTAACCATGGGCGAATACGGAGACGTGTCGCTGATGTGCAGAGTCGCTAGTGGCGTCGATCTGGCACAAACCGTTGTTCTTGAGCTCGACAAGACAGCAGAACACCTCCCCACGGCATGGCAGGTTCACAGAGATTGGTTTAACGATCTAGCTCTGCCTTGGAAACATGAGGGAATGGTGGGTTGGAACAATGCTGAACGCCTGGTTGAGTTTGGAGTTCCTCATGCCGTGAAGATGGATGTT\"], [447, \"1E\", \"TCACTGTGGACTGTTTGGGAAGGGCAGTATTGTCACCTGTGTCAAGGCATCGTGTGAGGCAAAAAAGAAAGCCACAGGACATGTGTACGACGCCCACAAGATAGTGTACACAGTCAAGGTTGAGCCACACACAGGAAACTATGTCGCGGCCAATGAAACCCACAGTGGGAGGAAAACCGCATTGTTCACAGTCTCTTCGGAAAAAACAATTCTAACCATGGGCGAATACGGAGACGTGTCGCTGATGTGCAGAGTCGCTAGTGGCGTTGATCTGGCACAAACCGTTGTTCTTGAGCTCGACAAGACAGCAGAACACCTCCCCACGGCATGGCAGGTTCACAGAGATTGGTTTAACGATCTAGCTCTGCCTTGGAAACATGAGGGAATGGTGGGTTGGAACAATGCTGAACGCCTGGTTGAGTTTGGAGTTCCTCATGCCGTGAAGATGGATGTT\"], [446, \"1C\", \"TCACTGTGGACTGTTTGGGAAGGGCAGTATTGTCACCTGTGTCAAGGCATCGTGTGAGGCAAGAAAGAAAGCCACAGGACATGTGTACGATGCCAACAAGATAGTGTACACAGTCAAGGTTGAGCCACACACAGGAAACTATGTCGCGGCCAATGAAACCCACAGTGGGAGGAAAACCGCATTGTTCACAGTCTCTTCGGAAAAAACAATTCTAACCATGGGCGAATACGGAGACGTGTCGCTGATGTGCAGAGTCGCTAGTGGCGTTGATCTGGCACAAACCGTTGTTCTTGAGCTCGACAAGACAGCAGAACACCTCCCCACGGCATGGCAGGTTCACAGAGATTGGTTTAACGATCTAGCTCTGCCTTGGAAACATGAGGGAATGGTGGGTTGGAACAATGCTGAACGCCTGGTTGAGTTTGGAGTTCCTCATGCCGTGAAGATGGATGTT\"], [442, \"1D\", \"TCACTGTGGACTGTTTGGGAAAGGCAGCATTGTTACCTGTGTCAAGGCATCGTGTGAGGCAAAGAAGAAAGCCACAGGATATGTGTATGACGCTAACAAGATAGTGTACACAGTCAAGGTTGAGCCACATACAGGAAACTATGTCGCGGCCAATGAAACCCATAGTGGGAGAAAAACCGCATTGTTCACAGTCTCTTCGGAAAAAACAATTCTAACCATGGGCGAATACGGAGACGTGTCGCTGATGTGCAGAGTCGCTAGTGGCGTCGATCTGGCACAAACCGTTGTTCTTGAGCTCGACAAGACAGCAGAACACCTCCCCACGGCATGGCAGGTTCACAGAGATTGGTTTAACGATCTAGCTCTGCCTTGGAAACATGAGGGAATGGTGGGTTGGAACAATGCTGAACGCCTGGTTGAGTTTGGAGTTCCTCACGCCGTGAAGATGGATGTT\"], [411, \"3A\", \"TCATTGTGGACTGTTCGGGAAGGGCAGTATTGTTACCTGTGTTAAGGCATCATGTGAGGCAAAAAAGAAAGCCACGGGACACGTGTATGATGCAAACAAGATAGTGTACACAGTCAAGGTCGAGCCACACACAGGAAACTATGTTGCAGCCAATGAAACCCACAGTGGGAGGAAAACCGCGTTATTTACGGTCTCTTCGGAAAAAACCATCTTGACCATGGGTGAATATGGAGACGTGTCATTGATGTGCAGAGTTGCCAGCGGCGTTGACTTGGCACAGACCGTTGTTCTTGAGCTCGACAAGACAGCAGAACACCTCCCTACGGCATGGCAGGTTCACAGAGACTGGTTTAATGACCTGGCTCTGCCATGGAAGCATGAGGGAATGGTGGGTTGGAACAATGCAGAACGCCTGGTTGAGTTCGGAGTTCCTCATGCCGTGAAGATGGACGTG\"], [408, \"3F\", \"TCATTGTGGACTGTTCGGGAAGGGCAGTATTGTTACCTGTGTTAAGGCATCATGTGAGGCAAAAAAGAAAGCCACGGGATACGTGTATGATGCAAACAAGATAGTGTATACAGTCAAGGTCGAGCCACACACAGGAAACTATGTTGCAGCCAATGAAACCCACAGTGGGAGGAAAACCGCGTTATTCACGGTCTCTTCGGAAAAGACCATCTTGACCATGGGTGAATATGGAGACGTGTCATTGATGTGCAGAGTTGCCAGTGGCGTTGACTTGGCACAGACCGTTGTTCTTGAGCTCGACAAGACAGCAGAACACCTCCCTACGGCATGGCAGGTTCACAGAGACTGGTTTAATGACCTGGCTCTGCCATGGAAGCATGAGGGAATGATGGGATGGAACAATGCAGAACGCCTGGTTGAGTTCGGAGTTCCTCATGCCGTGAAGATGGACGTG\"], [404, \"2D\", \"TCATTGCGGATTGTTTGGGAAAGGTAGCATTGTCACCTGTGTCAAGGCATCATGTGAGGCAAAAAAGAAAGCCACGGGATATGTTTATGATGCAAACAAGATAGTGTACACGGTCAAGGTTGAGCCACACACAGGGAACTATGTCGCAGCCAACGAAACCCACAGTGGGAGGAAAACTGCGCTGTTCACGGTCTCCTCGGAAAAAACCATCCTGACCATGGGTGAATATGGAGACGTGTCATTGATGTGTAGAGTCGCTAGTGGCGTTGACTTGGCACAGACCGTTGTCCTCGAGCTTGACAAGACAGCAGAACACCTTCCCACAGCATGGCAGGTTCACAGAGATTGGTTTAATGATCTGGCTCTGCCATGGAAACATGAGGGAATGGTGGGCTGGAACAATGCAGAACGCCTGGTTGAGTTTGGAGTCCCGCATGCTGTGAAGATGGACGTT\"]]", "scores_protein": "[[-1, \"Test\", \"HCGLFGKGSIVTCVKASCEAKKKATGHVYDANKIVYTVKVEPHTGNYVAANETHSGRKTALFTVSSEKTILTMGDYGDVSLMCRVASGVDLAQTVVLELDKTAEHLPTAWQVHRDWFNDLALPWKHEGMVGWNNAERLVEFGVPHAVKMDV\"], [151, \"1B\", \"HCGLFGKGSIVTCVKASCEAKKKATGHVYDANKIVYTVKVEPHTGNYVAANETHSGRKTALFTVSSEKTILTMGDYGDVSLMCRVASGVDLAQTVVLELDKTAEHLPTAWQVHRDWFNDLALPWKHEGMVGWNNAERLVEFGVPHAVKMDV\"], [150, \"1A\", \"HCGLFGKGSIVTCVKASCEAKKKATGHVYDANKIVYTVKVEPHTGNYVAANETHSGRKTALFTVSSEKTILTMGEYGDVSLMCRVASGVDLAQTVVLELDKTAEHLPTAWQVHRDWFNDLALPWKHEGMVGWNNAERLVEFGVPHAVKMDV\"], [150, \"3A\", \"HCGLFGKGSIVTCVKASCEAKKKATGHVYDANKIVYTVKVEPHTGNYVAANETHSGRKTALFTVSSEKTILTMGEYGDVSLMCRVASGVDLAQTVVLELDKTAEHLPTAWQVHRDWFNDLALPWKHEGMVGWNNAERLVEFGVPHAVKMDV\"], [149, \"1C\", \"HCGLFGKGSIVTCVKASCEARKKATGHVYDANKIVYTVKVEPHTGNYVAANETHSGRKTALFTVSSEKTILTMGEYGDVSLMCRVASGVDLAQTVVLELDKTAEHLPTAWQVHRDWFNDLALPWKHEGMVGWNNAERLVEFGVPHAVKMDV\"], [149, \"1D\", \"HCGLFGKGSIVTCVKASCEAKKKATGYVYDANKIVYTVKVEPHTGNYVAANETHSGRKTALFTVSSEKTILTMGEYGDVSLMCRVASGVDLAQTVVLELDKTAEHLPTAWQVHRDWFNDLALPWKHEGMVGWNNAERLVEFGVPHAVKMDV\"], [149, \"2D\", \"HCGLFGKGSIVTCVKASCEAKKKATGYVYDANKIVYTVKVEPHTGNYVAANETHSGRKTALFTVSSEKTILTMGEYGDVSLMCRVASGVDLAQTVVLELDKTAEHLPTAWQVHRDWFNDLALPWKHEGMVGWNNAERLVEFGVPHAVKMDV\"], [149, \"1E\", \"HCGLFGKGSIVTCVKASCEAKKKATGHVYDAHKIVYTVKVEPHTGNYVAANETHSGRKTALFTVSSEKTILTMGEYGDVSLMCRVASGVDLAQTVVLELDKTAEHLPTAWQVHRDWFNDLALPWKHEGMVGWNNAERLVEFGVPHAVKMDV\"], [148, \"3F\", \"HCGLFGKGSIVTCVKASCEAKKKATGYVYDANKIVYTVKVEPHTGNYVAANETHSGRKTALFTVSSEKTILTMGEYGDVSLMCRVASGVDLAQTVVLELDKTAEHLPTAWQVHRDWFNDLALPWKHEGMMGWNNAERLVEFGVPHAVKMDV\"]]", "subtype": "OHFV-1", "tree": "((((Test:0.00000,1B:0.00000):0.00835,(1C:0.00332,1E:0.00332):0.00503):0.00254,(1A:0.00666,1D:0.00666):0.00423):0.04498,((3A:0.00779,3F:0.00779):0.03995,2D:0.04773):0.00813);" }, "sequence": "TCACTGTGGACTGTTTGGGAAGGGCAGTATTGTCACCTGTGTCAAGGCATCGTGTGAGGCAAAAAAGAAAGCCACAGGACACGTGTACGACGCTAACAAGATAGTGTACACAGTCAAGGTTGAGCCACATACAGGAAACTATGTCGCGGCCAATGAAACCCACAGTGGGAGGAAAACCGCATTGTTCACAGTCTCTTCGGAAAAAACAATTCTAACCATGGGCGATTACGGGGACGTGTCGCTGATGTGCAGAGTCGCTAGTGGCGTCGATCTGGCACAAACCGTTGTTCTTGAGCTCGACAAGACAGCAGAACACCTCCCCACGGCATGGCAGGTTCACAGAGATTGGTTTAACGATCTAGCTCTGCCTTGGAAACATGAGGGAATGGTGGGTTGGAACAATGCTGAACGCCTGGTTGAGTTTGGAGTTCCTCATGCCGTGAAGATGGATGTT", "status": "successful" } ], "type": "api" } -
To access the details of a task, you should trigger an HTTP request to
/api/taskswith these parameters:key: Your valid API key. (required)pid: The ID of the project. (required)tid: The ID of the task. (required)
And the response for a successful task will be something like this:https://ohfv.viroinformatics.com/api/tasks?key=YOUR_API_KEY&pid=05f5b4d4804f47318907087827f7eb7f&tid=50ec97de93b64000a47a9de295768668{ "datetime": { "end": "2022-03-09 14:11:04.603747", "start": "2022-03-09 14:10:59.000907" }, "id": "50ec97de93b64000a47a9de295768668", "log": [ [ "message", "2022-03-09 14:10:59.034277", "Analysis started." ], [ "message", "2022-03-09 14:10:59.066561", "The initial validity of the sequence is confirmed." ], [ "message", "2022-03-09 14:10:59.144560", "FASTA file including query and prototypes strains is created." ], [ "message", "2022-03-09 14:10:59.425692", "Determining and validating the target fragment is started." ], [ "message", "2022-03-09 14:10:59.426327", "The sequence is successfully determined and verified." ], [ "message", "2022-03-09 14:10:59.428535", "Aligned FASTA file including query and prototypes is created." ], [ "message", "2022-03-09 14:11:04.448737", "Alignment scores are computed." ], [ "message", "2022-03-09 14:11:04.449943", "Necessary files for constructing phylogenetic tree are created." ], [ "message", "2022-03-09 14:11:04.596387", "Phylogenetic tree is successfully constructed." ], [ "message", "2022-03-09 14:11:04.597070", "The clusteron of the sequence is successfully determined." ], [ "success", "2022-03-09 14:11:04.597640", "Analysis completed successfully." ] ], "name": "Test", "report": { "aligned": ">Test\nTCACTGTGGACTGTTTGGGAAGGGCAGTATTGTCACCTGTGTCAAGGCATCGTGTGAGGC\nAAAAAAGAAAGCCACAGGACACGTGTACGACGCTAACAAGATAGTGTACACAGTCAAGGT\nTGAGCCACATACAGGAAACTATGTCGCGGCCAATGAAACCCACAGTGGGAGGAAAACCGC\nATTGTTCACAGTCTCTTCGGAAAAAACAATTCTAACCATGGGCGATTACGGGGACGTGTC\nGCTGATGTGCAGAGTCGCTAGTGGCGTCGATCTGGCACAAACCGTTGTTCTTGAGCTCGA\nCAAGACAGCAGAACACCTCCCCACGGCATGGCAGGTTCACAGAGATTGGTTTAACGATCT\nAGCTCTGCCTTGGAAACATGAGGGAATGGTGGGTTGGAACAATGCTGAACGCCTGGTTGA\nGTTTGGAGTTCCTCATGCCGTGAAGATGGATGTT\n>1A\nTCACTGTGGACTGTTTGGGAAGGGCAGTATTGTCACCTGTGTCAAGGCATCGTGTGAGGC\nAAAGAAGAAAGCCACAGGACATGTGTATGACGCTAACAAGATAGTGTACACAGTCAAGGT\nTGAGCCACATACAGGAAACTATGTCGCGGCCAATGAAACCCATAGTGGGAGGAAAACCGC\nATTGTTCACAGTCTCTTCGGAAAAAACAATTCTAACCATGGGCGAATACGGAGACGTGTC\nGCTGATGTGCAGAGTCGCTAGTGGCGTCGATCTGGCACAAACCGTTGTTCTTGAGCTCGA\nCAAGACAGCAGAACACCTCCCCACGGCATGGCAGGTTCACAGAGATTGGTTTAACGATCT\nAGCTCTGCCTTGGAAACATGAGGGAATGGTGGGTTGGAACAATGCTGAACGCCTGGTTGA\nGTTTGGAGTTCCTCATGCCGTGAAGATGGATGTT\n>3A\nTCATTGTGGACTGTTCGGGAAGGGCAGTATTGTTACCTGTGTTAAGGCATCATGTGAGGC\nAAAAAAGAAAGCCACGGGACACGTGTATGATGCAAACAAGATAGTGTACACAGTCAAGGT\nCGAGCCACACACAGGAAACTATGTTGCAGCCAATGAAACCCACAGTGGGAGGAAAACCGC\nGTTATTTACGGTCTCTTCGGAAAAAACCATCTTGACCATGGGTGAATATGGAGACGTGTC\nATTGATGTGCAGAGTTGCCAGCGGCGTTGACTTGGCACAGACCGTTGTTCTTGAGCTCGA\nCAAGACAGCAGAACACCTCCCTACGGCATGGCAGGTTCACAGAGACTGGTTTAATGACCT\nGGCTCTGCCATGGAAGCATGAGGGAATGGTGGGTTGGAACAATGCAGAACGCCTGGTTGA\nGTTCGGAGTTCCTCATGCCGTGAAGATGGACGTG\n>1B\nTCACTGTGGACTGTTTGGGAAGGGCAGTATTGTCACCTGTGTCAAGGCATCGTGTGAGGC\nAAAAAAGAAAGCCACAGGACACGTGTACGACGCTAACAAGATAGTGTACACAGTCAAGGT\nTGAGCCACATACAGGAAACTATGTCGCGGCCAATGAAACCCACAGTGGGAGGAAAACCGC\nATTGTTCACAGTCTCTTCGGAAAAAACAATTCTAACCATGGGCGATTACGGGGACGTGTC\nGCTGATGTGCAGAGTCGCTAGTGGCGTCGATCTGGCACAAACCGTTGTTCTTGAGCTCGA\nCAAGACAGCAGAACACCTCCCCACGGCATGGCAGGTTCACAGAGATTGGTTTAACGATCT\nAGCTCTGCCTTGGAAACATGAGGGAATGGTGGGTTGGAACAATGCTGAACGCCTGGTTGA\nGTTTGGAGTTCCTCATGCCGTGAAGATGGATGTT\n>1C\nTCACTGTGGACTGTTTGGGAAGGGCAGTATTGTCACCTGTGTCAAGGCATCGTGTGAGGC\nAAGAAAGAAAGCCACAGGACATGTGTACGATGCCAACAAGATAGTGTACACAGTCAAGGT\nTGAGCCACACACAGGAAACTATGTCGCGGCCAATGAAACCCACAGTGGGAGGAAAACCGC\nATTGTTCACAGTCTCTTCGGAAAAAACAATTCTAACCATGGGCGAATACGGAGACGTGTC\nGCTGATGTGCAGAGTCGCTAGTGGCGTTGATCTGGCACAAACCGTTGTTCTTGAGCTCGA\nCAAGACAGCAGAACACCTCCCCACGGCATGGCAGGTTCACAGAGATTGGTTTAACGATCT\nAGCTCTGCCTTGGAAACATGAGGGAATGGTGGGTTGGAACAATGCTGAACGCCTGGTTGA\nGTTTGGAGTTCCTCATGCCGTGAAGATGGATGTT\n>1D\nTCACTGTGGACTGTTTGGGAAAGGCAGCATTGTTACCTGTGTCAAGGCATCGTGTGAGGC\nAAAGAAGAAAGCCACAGGATATGTGTATGACGCTAACAAGATAGTGTACACAGTCAAGGT\nTGAGCCACATACAGGAAACTATGTCGCGGCCAATGAAACCCATAGTGGGAGAAAAACCGC\nATTGTTCACAGTCTCTTCGGAAAAAACAATTCTAACCATGGGCGAATACGGAGACGTGTC\nGCTGATGTGCAGAGTCGCTAGTGGCGTCGATCTGGCACAAACCGTTGTTCTTGAGCTCGA\nCAAGACAGCAGAACACCTCCCCACGGCATGGCAGGTTCACAGAGATTGGTTTAACGATCT\nAGCTCTGCCTTGGAAACATGAGGGAATGGTGGGTTGGAACAATGCTGAACGCCTGGTTGA\nGTTTGGAGTTCCTCACGCCGTGAAGATGGATGTT\n>2D\nTCATTGCGGATTGTTTGGGAAAGGTAGCATTGTCACCTGTGTCAAGGCATCATGTGAGGC\nAAAAAAGAAAGCCACGGGATATGTTTATGATGCAAACAAGATAGTGTACACGGTCAAGGT\nTGAGCCACACACAGGGAACTATGTCGCAGCCAACGAAACCCACAGTGGGAGGAAAACTGC\nGCTGTTCACGGTCTCCTCGGAAAAAACCATCCTGACCATGGGTGAATATGGAGACGTGTC\nATTGATGTGTAGAGTCGCTAGTGGCGTTGACTTGGCACAGACCGTTGTCCTCGAGCTTGA\nCAAGACAGCAGAACACCTTCCCACAGCATGGCAGGTTCACAGAGATTGGTTTAATGATCT\nGGCTCTGCCATGGAAACATGAGGGAATGGTGGGCTGGAACAATGCAGAACGCCTGGTTGA\nGTTTGGAGTCCCGCATGCTGTGAAGATGGACGTT\n>1E\nTCACTGTGGACTGTTTGGGAAGGGCAGTATTGTCACCTGTGTCAAGGCATCGTGTGAGGC\nAAAAAAGAAAGCCACAGGACATGTGTACGACGCCCACAAGATAGTGTACACAGTCAAGGT\nTGAGCCACACACAGGAAACTATGTCGCGGCCAATGAAACCCACAGTGGGAGGAAAACCGC\nATTGTTCACAGTCTCTTCGGAAAAAACAATTCTAACCATGGGCGAATACGGAGACGTGTC\nGCTGATGTGCAGAGTCGCTAGTGGCGTTGATCTGGCACAAACCGTTGTTCTTGAGCTCGA\nCAAGACAGCAGAACACCTCCCCACGGCATGGCAGGTTCACAGAGATTGGTTTAACGATCT\nAGCTCTGCCTTGGAAACATGAGGGAATGGTGGGTTGGAACAATGCTGAACGCCTGGTTGA\nGTTTGGAGTTCCTCATGCCGTGAAGATGGATGTT\n>3F\nTCATTGTGGACTGTTCGGGAAGGGCAGTATTGTTACCTGTGTTAAGGCATCATGTGAGGC\nAAAAAAGAAAGCCACGGGATACGTGTATGATGCAAACAAGATAGTGTATACAGTCAAGGT\nCGAGCCACACACAGGAAACTATGTTGCAGCCAATGAAACCCACAGTGGGAGGAAAACCGC\nGTTATTCACGGTCTCTTCGGAAAAGACCATCTTGACCATGGGTGAATATGGAGACGTGTC\nATTGATGTGCAGAGTTGCCAGTGGCGTTGACTTGGCACAGACCGTTGTTCTTGAGCTCGA\nCAAGACAGCAGAACACCTCCCTACGGCATGGCAGGTTCACAGAGACTGGTTTAATGACCT\nGGCTCTGCCATGGAAGCATGAGGGAATGATGGGATGGAACAATGCAGAACGCCTGGTTGA\nGTTCGGAGTTCCTCATGCCGTGAAGATGGACGTG\n", "clusteron": "1B", "lineage": "None", "scores_dna": "[[-1, \"Test\", \"TCACTGTGGACTGTTTGGGAAGGGCAGTATTGTCACCTGTGTCAAGGCATCGTGTGAGGCAAAAAAGAAAGCCACAGGACACGTGTACGACGCTAACAAGATAGTGTACACAGTCAAGGTTGAGCCACATACAGGAAACTATGTCGCGGCCAATGAAACCCACAGTGGGAGGAAAACCGCATTGTTCACAGTCTCTTCGGAAAAAACAATTCTAACCATGGGCGATTACGGGGACGTGTCGCTGATGTGCAGAGTCGCTAGTGGCGTCGATCTGGCACAAACCGTTGTTCTTGAGCTCGACAAGACAGCAGAACACCTCCCCACGGCATGGCAGGTTCACAGAGATTGGTTTAACGATCTAGCTCTGCCTTGGAAACATGAGGGAATGGTGGGTTGGAACAATGCTGAACGCCTGGTTGAGTTTGGAGTTCCTCATGCCGTGAAGATGGATGTT\"], [454, \"1B\", \"TCACTGTGGACTGTTTGGGAAGGGCAGTATTGTCACCTGTGTCAAGGCATCGTGTGAGGCAAAAAAGAAAGCCACAGGACACGTGTACGACGCTAACAAGATAGTGTACACAGTCAAGGTTGAGCCACATACAGGAAACTATGTCGCGGCCAATGAAACCCACAGTGGGAGGAAAACCGCATTGTTCACAGTCTCTTCGGAAAAAACAATTCTAACCATGGGCGATTACGGGGACGTGTCGCTGATGTGCAGAGTCGCTAGTGGCGTCGATCTGGCACAAACCGTTGTTCTTGAGCTCGACAAGACAGCAGAACACCTCCCCACGGCATGGCAGGTTCACAGAGATTGGTTTAACGATCTAGCTCTGCCTTGGAAACATGAGGGAATGGTGGGTTGGAACAATGCTGAACGCCTGGTTGAGTTTGGAGTTCCTCATGCCGTGAAGATGGATGTT\"], [448, \"1A\", \"TCACTGTGGACTGTTTGGGAAGGGCAGTATTGTCACCTGTGTCAAGGCATCGTGTGAGGCAAAGAAGAAAGCCACAGGACATGTGTATGACGCTAACAAGATAGTGTACACAGTCAAGGTTGAGCCACATACAGGAAACTATGTCGCGGCCAATGAAACCCATAGTGGGAGGAAAACCGCATTGTTCACAGTCTCTTCGGAAAAAACAATTCTAACCATGGGCGAATACGGAGACGTGTCGCTGATGTGCAGAGTCGCTAGTGGCGTCGATCTGGCACAAACCGTTGTTCTTGAGCTCGACAAGACAGCAGAACACCTCCCCACGGCATGGCAGGTTCACAGAGATTGGTTTAACGATCTAGCTCTGCCTTGGAAACATGAGGGAATGGTGGGTTGGAACAATGCTGAACGCCTGGTTGAGTTTGGAGTTCCTCATGCCGTGAAGATGGATGTT\"], [447, \"1E\", \"TCACTGTGGACTGTTTGGGAAGGGCAGTATTGTCACCTGTGTCAAGGCATCGTGTGAGGCAAAAAAGAAAGCCACAGGACATGTGTACGACGCCCACAAGATAGTGTACACAGTCAAGGTTGAGCCACACACAGGAAACTATGTCGCGGCCAATGAAACCCACAGTGGGAGGAAAACCGCATTGTTCACAGTCTCTTCGGAAAAAACAATTCTAACCATGGGCGAATACGGAGACGTGTCGCTGATGTGCAGAGTCGCTAGTGGCGTTGATCTGGCACAAACCGTTGTTCTTGAGCTCGACAAGACAGCAGAACACCTCCCCACGGCATGGCAGGTTCACAGAGATTGGTTTAACGATCTAGCTCTGCCTTGGAAACATGAGGGAATGGTGGGTTGGAACAATGCTGAACGCCTGGTTGAGTTTGGAGTTCCTCATGCCGTGAAGATGGATGTT\"], [446, \"1C\", \"TCACTGTGGACTGTTTGGGAAGGGCAGTATTGTCACCTGTGTCAAGGCATCGTGTGAGGCAAGAAAGAAAGCCACAGGACATGTGTACGATGCCAACAAGATAGTGTACACAGTCAAGGTTGAGCCACACACAGGAAACTATGTCGCGGCCAATGAAACCCACAGTGGGAGGAAAACCGCATTGTTCACAGTCTCTTCGGAAAAAACAATTCTAACCATGGGCGAATACGGAGACGTGTCGCTGATGTGCAGAGTCGCTAGTGGCGTTGATCTGGCACAAACCGTTGTTCTTGAGCTCGACAAGACAGCAGAACACCTCCCCACGGCATGGCAGGTTCACAGAGATTGGTTTAACGATCTAGCTCTGCCTTGGAAACATGAGGGAATGGTGGGTTGGAACAATGCTGAACGCCTGGTTGAGTTTGGAGTTCCTCATGCCGTGAAGATGGATGTT\"], [442, \"1D\", \"TCACTGTGGACTGTTTGGGAAAGGCAGCATTGTTACCTGTGTCAAGGCATCGTGTGAGGCAAAGAAGAAAGCCACAGGATATGTGTATGACGCTAACAAGATAGTGTACACAGTCAAGGTTGAGCCACATACAGGAAACTATGTCGCGGCCAATGAAACCCATAGTGGGAGAAAAACCGCATTGTTCACAGTCTCTTCGGAAAAAACAATTCTAACCATGGGCGAATACGGAGACGTGTCGCTGATGTGCAGAGTCGCTAGTGGCGTCGATCTGGCACAAACCGTTGTTCTTGAGCTCGACAAGACAGCAGAACACCTCCCCACGGCATGGCAGGTTCACAGAGATTGGTTTAACGATCTAGCTCTGCCTTGGAAACATGAGGGAATGGTGGGTTGGAACAATGCTGAACGCCTGGTTGAGTTTGGAGTTCCTCACGCCGTGAAGATGGATGTT\"], [411, \"3A\", \"TCATTGTGGACTGTTCGGGAAGGGCAGTATTGTTACCTGTGTTAAGGCATCATGTGAGGCAAAAAAGAAAGCCACGGGACACGTGTATGATGCAAACAAGATAGTGTACACAGTCAAGGTCGAGCCACACACAGGAAACTATGTTGCAGCCAATGAAACCCACAGTGGGAGGAAAACCGCGTTATTTACGGTCTCTTCGGAAAAAACCATCTTGACCATGGGTGAATATGGAGACGTGTCATTGATGTGCAGAGTTGCCAGCGGCGTTGACTTGGCACAGACCGTTGTTCTTGAGCTCGACAAGACAGCAGAACACCTCCCTACGGCATGGCAGGTTCACAGAGACTGGTTTAATGACCTGGCTCTGCCATGGAAGCATGAGGGAATGGTGGGTTGGAACAATGCAGAACGCCTGGTTGAGTTCGGAGTTCCTCATGCCGTGAAGATGGACGTG\"], [408, \"3F\", \"TCATTGTGGACTGTTCGGGAAGGGCAGTATTGTTACCTGTGTTAAGGCATCATGTGAGGCAAAAAAGAAAGCCACGGGATACGTGTATGATGCAAACAAGATAGTGTATACAGTCAAGGTCGAGCCACACACAGGAAACTATGTTGCAGCCAATGAAACCCACAGTGGGAGGAAAACCGCGTTATTCACGGTCTCTTCGGAAAAGACCATCTTGACCATGGGTGAATATGGAGACGTGTCATTGATGTGCAGAGTTGCCAGTGGCGTTGACTTGGCACAGACCGTTGTTCTTGAGCTCGACAAGACAGCAGAACACCTCCCTACGGCATGGCAGGTTCACAGAGACTGGTTTAATGACCTGGCTCTGCCATGGAAGCATGAGGGAATGATGGGATGGAACAATGCAGAACGCCTGGTTGAGTTCGGAGTTCCTCATGCCGTGAAGATGGACGTG\"], [404, \"2D\", \"TCATTGCGGATTGTTTGGGAAAGGTAGCATTGTCACCTGTGTCAAGGCATCATGTGAGGCAAAAAAGAAAGCCACGGGATATGTTTATGATGCAAACAAGATAGTGTACACGGTCAAGGTTGAGCCACACACAGGGAACTATGTCGCAGCCAACGAAACCCACAGTGGGAGGAAAACTGCGCTGTTCACGGTCTCCTCGGAAAAAACCATCCTGACCATGGGTGAATATGGAGACGTGTCATTGATGTGTAGAGTCGCTAGTGGCGTTGACTTGGCACAGACCGTTGTCCTCGAGCTTGACAAGACAGCAGAACACCTTCCCACAGCATGGCAGGTTCACAGAGATTGGTTTAATGATCTGGCTCTGCCATGGAAACATGAGGGAATGGTGGGCTGGAACAATGCAGAACGCCTGGTTGAGTTTGGAGTCCCGCATGCTGTGAAGATGGACGTT\"]]", "scores_protein": "[[-1, \"Test\", \"HCGLFGKGSIVTCVKASCEAKKKATGHVYDANKIVYTVKVEPHTGNYVAANETHSGRKTALFTVSSEKTILTMGDYGDVSLMCRVASGVDLAQTVVLELDKTAEHLPTAWQVHRDWFNDLALPWKHEGMVGWNNAERLVEFGVPHAVKMDV\"], [151, \"1B\", \"HCGLFGKGSIVTCVKASCEAKKKATGHVYDANKIVYTVKVEPHTGNYVAANETHSGRKTALFTVSSEKTILTMGDYGDVSLMCRVASGVDLAQTVVLELDKTAEHLPTAWQVHRDWFNDLALPWKHEGMVGWNNAERLVEFGVPHAVKMDV\"], [150, \"1A\", \"HCGLFGKGSIVTCVKASCEAKKKATGHVYDANKIVYTVKVEPHTGNYVAANETHSGRKTALFTVSSEKTILTMGEYGDVSLMCRVASGVDLAQTVVLELDKTAEHLPTAWQVHRDWFNDLALPWKHEGMVGWNNAERLVEFGVPHAVKMDV\"], [150, \"3A\", \"HCGLFGKGSIVTCVKASCEAKKKATGHVYDANKIVYTVKVEPHTGNYVAANETHSGRKTALFTVSSEKTILTMGEYGDVSLMCRVASGVDLAQTVVLELDKTAEHLPTAWQVHRDWFNDLALPWKHEGMVGWNNAERLVEFGVPHAVKMDV\"], [149, \"1C\", \"HCGLFGKGSIVTCVKASCEARKKATGHVYDANKIVYTVKVEPHTGNYVAANETHSGRKTALFTVSSEKTILTMGEYGDVSLMCRVASGVDLAQTVVLELDKTAEHLPTAWQVHRDWFNDLALPWKHEGMVGWNNAERLVEFGVPHAVKMDV\"], [149, \"1D\", \"HCGLFGKGSIVTCVKASCEAKKKATGYVYDANKIVYTVKVEPHTGNYVAANETHSGRKTALFTVSSEKTILTMGEYGDVSLMCRVASGVDLAQTVVLELDKTAEHLPTAWQVHRDWFNDLALPWKHEGMVGWNNAERLVEFGVPHAVKMDV\"], [149, \"2D\", \"HCGLFGKGSIVTCVKASCEAKKKATGYVYDANKIVYTVKVEPHTGNYVAANETHSGRKTALFTVSSEKTILTMGEYGDVSLMCRVASGVDLAQTVVLELDKTAEHLPTAWQVHRDWFNDLALPWKHEGMVGWNNAERLVEFGVPHAVKMDV\"], [149, \"1E\", \"HCGLFGKGSIVTCVKASCEAKKKATGHVYDAHKIVYTVKVEPHTGNYVAANETHSGRKTALFTVSSEKTILTMGEYGDVSLMCRVASGVDLAQTVVLELDKTAEHLPTAWQVHRDWFNDLALPWKHEGMVGWNNAERLVEFGVPHAVKMDV\"], [148, \"3F\", \"HCGLFGKGSIVTCVKASCEAKKKATGYVYDANKIVYTVKVEPHTGNYVAANETHSGRKTALFTVSSEKTILTMGEYGDVSLMCRVASGVDLAQTVVLELDKTAEHLPTAWQVHRDWFNDLALPWKHEGMMGWNNAERLVEFGVPHAVKMDV\"]]", "subtype": "OHFV-1", "tree": "((((Test:0.00000,1B:0.00000):0.00835,(1C:0.00332,1E:0.00332):0.00503):0.00254,(1A:0.00666,1D:0.00666):0.00423):0.04498,((3A:0.00779,3F:0.00779):0.03995,2D:0.04773):0.00813);" }, "sequence": "TCACTGTGGACTGTTTGGGAAGGGCAGTATTGTCACCTGTGTCAAGGCATCGTGTGAGGCAAAAAAGAAAGCCACAGGACACGTGTACGACGCTAACAAGATAGTGTACACAGTCAAGGTTGAGCCACATACAGGAAACTATGTCGCGGCCAATGAAACCCACAGTGGGAGGAAAACCGCATTGTTCACAGTCTCTTCGGAAAAAACAATTCTAACCATGGGCGATTACGGGGACGTGTCGCTGATGTGCAGAGTCGCTAGTGGCGTCGATCTGGCACAAACCGTTGTTCTTGAGCTCGACAAGACAGCAGAACACCTCCCCACGGCATGGCAGGTTCACAGAGATTGGTTTAACGATCTAGCTCTGCCTTGGAAACATGAGGGAATGGTGGGTTGGAACAATGCTGAACGCCTGGTTGAGTTTGGAGTTCCTCATGCCGTGAAGATGGATGTT", "status": "successful" }